CHLOROPLAST GENE EXPRESSION Transcription RNA processing splicing cleavages


- Slides: 23

CHLOROPLAST GENE EXPRESSION • Transcription • RNA processing (splicing, cleavages, modification) • Translation • Regulation • Dependence on nuclear genes

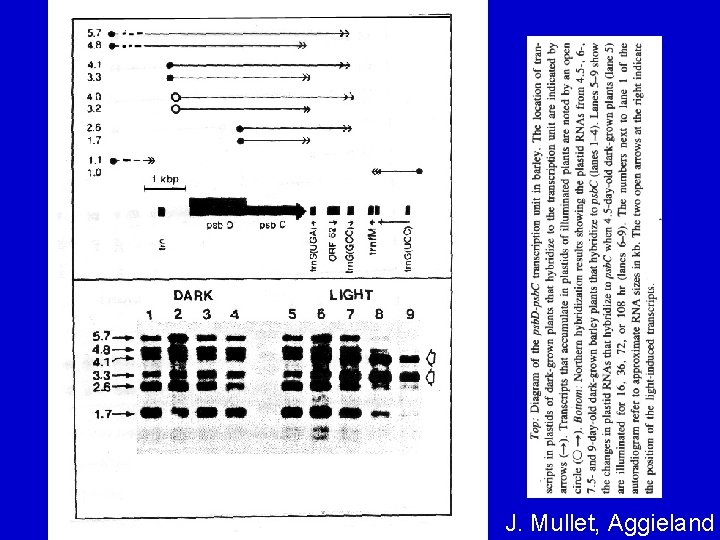
TRANSCRIPTION • Many, but not all, cp genes are arranged in operon-like units and co-transcribed – e. g. , psb. D-psb. C gene cluster (see next slide) – A unique feature of psb. D-psb. C gene transcription: a different (closer) promoter is used in the light called the light-responsive promoter (LRP).

J. Mullet, Aggieland

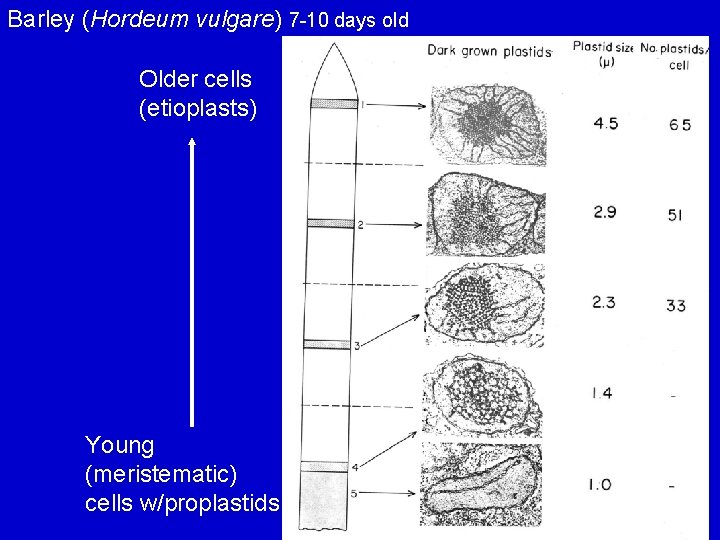
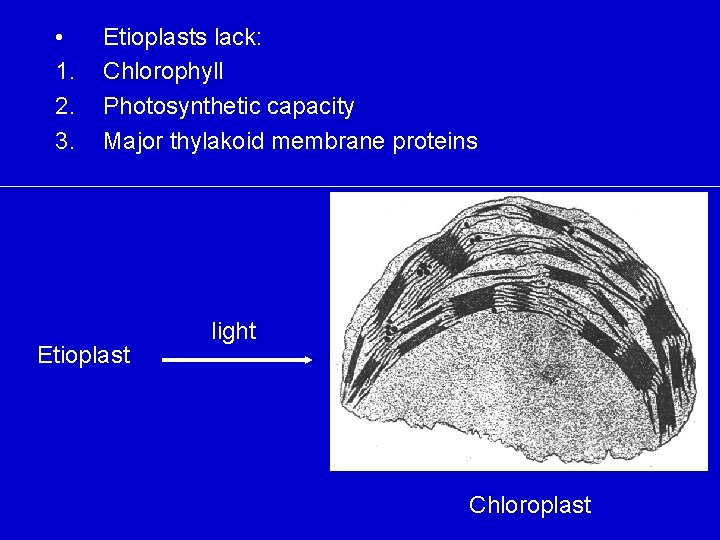
Barley (Hordeum vulgare) 7 -10 days old Older cells (etioplasts) Young (meristematic) cells w/proplastids

• 1. 2. 3. Etioplasts lack: Chlorophyll Photosynthetic capacity Major thylakoid membrane proteins Etioplast light Chloroplast

How many promoters in cp. DNA? • ~30 transcription units (promoters) in higher plant cp DNA – determined experimentally by capping of cp RNA with guanylyl transferase and radioactive GT 32 P, and hybridization to cp. DNA fragments. – The transferase attaches GMP to the 5’ end of RNAs that have 2 or 3 phosphates • Only primary transcription products have > 1 phosphate at the 5’ end of the RNA.

A "transcription unit" is determined by the position of the promoter (5') and terminator (3') signals. Terminators not clearly defined, but t. RNA genes seem to be good transcription terminators in chloroplasts.

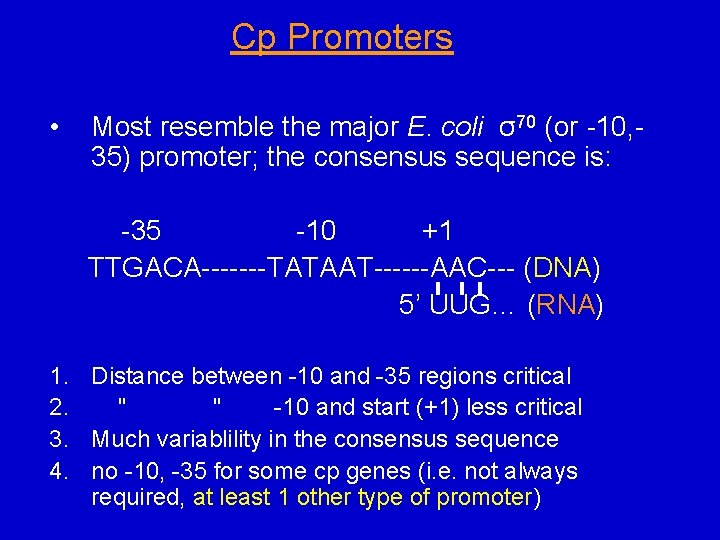
Cp Promoters • Most resemble the major E. coli σ70 (or -10, 35) promoter; the consensus sequence is: -35 -10 +1 TTGACA-------TATAAT------AAC--- (DNA) 5’ UUG… (RNA) 1. Distance between -10 and -35 regions critical 2. " " -10 and start (+1) less critical 3. Much variablility in the consensus sequence 4. no -10, -35 for some cp genes (i. e. not always required, at least 1 other type of promoter)

Control of Cp transcription • Transcription rate important : – mainly controlled at initiation step – determined in part by "promoter strength“ – also modulated for some genes (psb. D) by upstream sequences that bind regulatory proteins • Some genes have "alternative promoters" (e. g. , psb. D – psb. C) - also provides for regulation

CP RNA polymerases Two main forms in vascular plants: 1. E. coli or eubacterial-like polymerase (also called PEP, plastid-encoded polymerase) 2. Phage-like or NEP (nuclear-encoded polymerase) polymerase

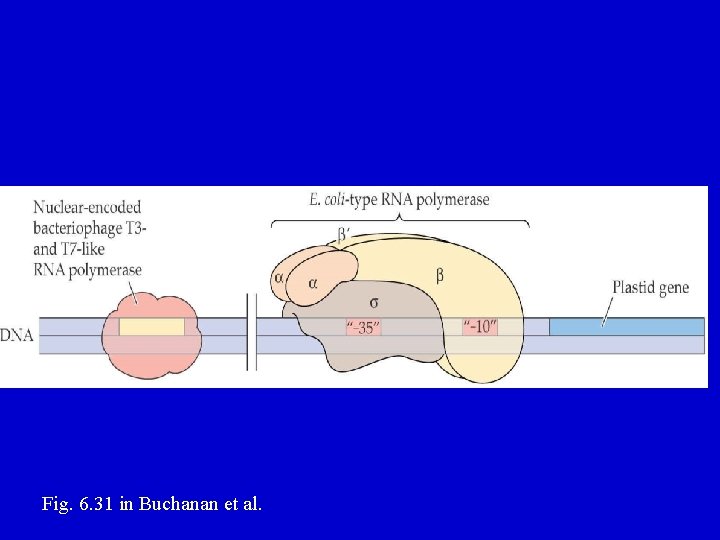
E. coli-like (PEP) polymerase • composed of Core + Sigma factor – Core = 4 subunits, α 2 ββ' • α is encoded by the rpo. A gene • β is encoded by the rpo. B gene • β' is encoded by the rpo. C 1 and rpo. C 2 genes – Sigma factor needed to initiate transcription at the bacterial promoter (recognizes -10, -35 regions) • Nuclear encoded, family of 6 genes in Arabidopsis • Inhibited by rifampicin

Fig. 6. 31 in Buchanan et al.

Phage-like (NEP) polymerase • Catalytic subunit is similar to the 1 -subunit phage (e. g. , T 7) and mitochondrial RNA polymerases • Nuclear gene • Enzyme insensitive to rifampicin • Promoter is usually a single region of 7 -10 bp (YRTA core), but other sequences stimulate • Evolution – Viral Origin? – Mitochondrial origin? – When did it get into plants?

Why two chloroplast RNA polymerases? NEP is more important early in plastid development when plastid transcription (and translation) is relatively low. - transcribes r. RNA, rpo and other genetic functions genes (GFG) PEP is more important in mature chloroplasts. - transcribes some GFG genes, but strongly transcribes photosynthesis genes

CP pre-m. RNA PROCESSING Most, if not all primary transcripts are processed by cleavage(s) or splicing or both CP m. RNAs are not polyadenylated, and are not "capped" (cap= 7 methylguanosine). • Nucleolytic Cleavages: 1. Endonucleases - cut internally (e. g. , between genes), fairly specific 2. Exonucleases - trim at 3' or 5'-ends, processive, less specific

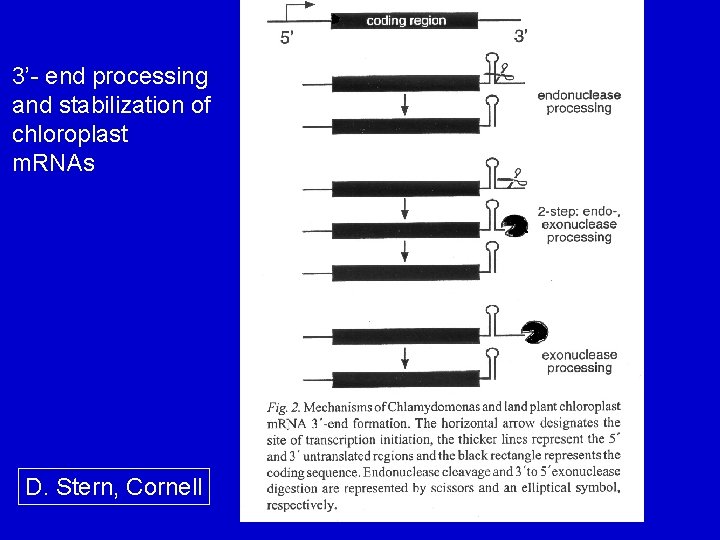
Inverted repeats in cp. RNA processing Inverted repeats occur at 3'-end of most cp protein-encoding genes. - processing sites, determine the 3'-end of m. RNAs - mechanisms: 1. proteins recognize the 3'-IR, bind and stop a processive exonuclease 2. An endonuclease cleaves at the 3’-IR 3. Combination of the two above

3’- end processing and stabilization of chloroplast m. RNAs D. Stern, Cornell

Pathways of Cp pre-m. RNA Processing & Degradation in Chlamydomonas (a) and (b) may use some of the same enzymes D. Stern, Cornell

Translation in Chloroplasts Translation machinery is bacteria-like: • Ribosomes: -70 S (composed of L (50 S) and S (30 S) subunits) -contain 23 S (L), 16 S (S), and 5 S (L) r. RNAs -each subunit (L and S) contains ~30 proteins • Initiation factors: if 1, if 2, if 3 • Elongation factors: ef-Tu, ef-Ts, and G • Translation is initiated with fmet (formylated Met) Chloroplast polyribosomes will use E. coli soluble factors for elongation and termination phases.

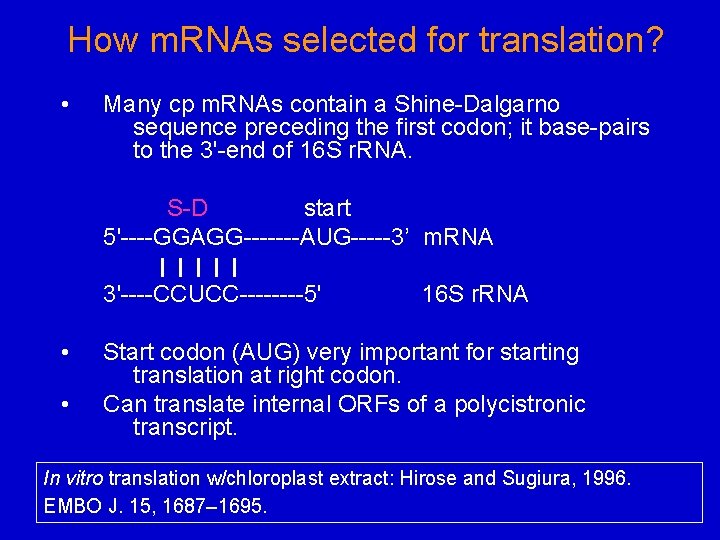
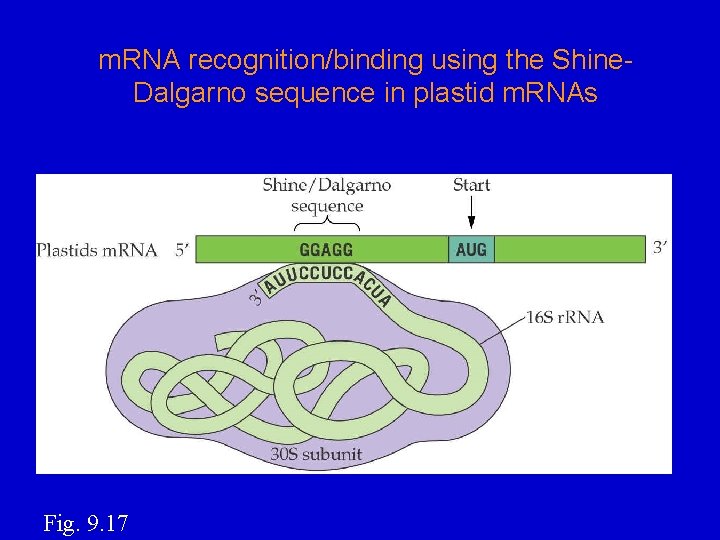
How m. RNAs selected for translation? • Many cp m. RNAs contain a Shine-Dalgarno sequence preceding the first codon; it base-pairs to the 3'-end of 16 S r. RNA. S-D start 5'----GGAGG-------AUG-----3’ m. RNA 3'----CCUCC----5' • • 16 S r. RNA Start codon (AUG) very important for starting translation at right codon. Can translate internal ORFs of a polycistronic transcript. In vitro translation w/chloroplast extract: Hirose and Sugiura, 1996. EMBO J. 15, 1687– 1695.

m. RNA recognition/binding using the Shine. Dalgarno sequence in plastid m. RNAs Fig. 9. 17

Differences with bacteria 1. Many chloroplast m. RNAs have relatively long (~ 300 nt) 5' untranslated regions (UTR) that bind proteins. 2. Many chloroplast m. RNAs don’t have a S-D sequence, and in 1 case, it suppresses translation (Sugiura lab). 3. Must be another initiation mechanism • • Scanning ? Some of the proteins that bind the 5’ UTRs of m. RNAs promote translation

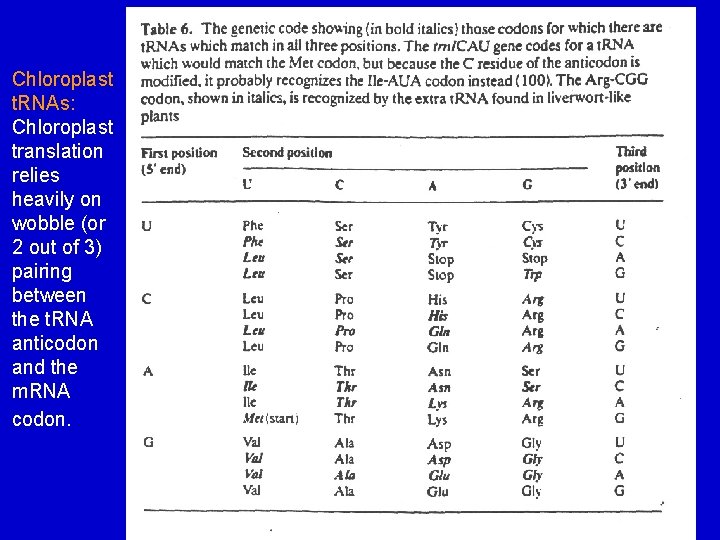
Chloroplast t. RNAs: Chloroplast translation relies heavily on wobble (or 2 out of 3) pairing between the t. RNA anticodon and the m. RNA codon.