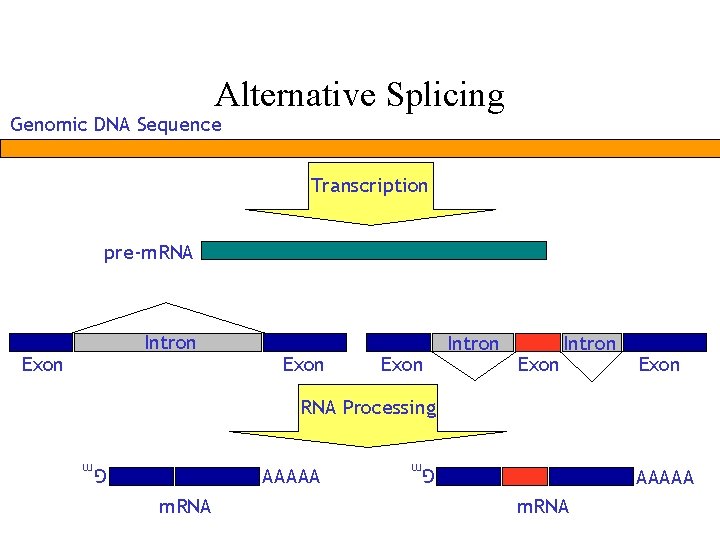
Alternative Splicing Genomic DNA Sequence Transcription prem RNA

- Slides: 25

Alternative Splicing Genomic DNA Sequence Transcription pre-m. RNA Intron Exon Intron Exon RNA Processing Gm m. RNA Gm AAAAA m. RNA

Alternative Splicing Data Sources are Large and Growing Curated databases SWISS-PROT and Ref. Seq both support annotation of experimentally supported alternative splicing c. DNA Sequencing Projects RIKEN sequenced >21000 full length mouse c. DNAs Many other projects underway (human, fly, plants, …) Shinagawa et al. (2001) Nature 409: 685 -90 Microarray detection Direct or indirect alternative splicing detection Hu et al. (2001)Genome Res 11: 1237 -45 Yeakley et al. (2002) Nat Biotech 20: 353 -9 Public EST data sources (db. EST) >4. 5 million human EST sequences >12 million total EST sequences About 1000 new sequences per day Boguski et al. (1993) Nat Gen 4: 332 -3

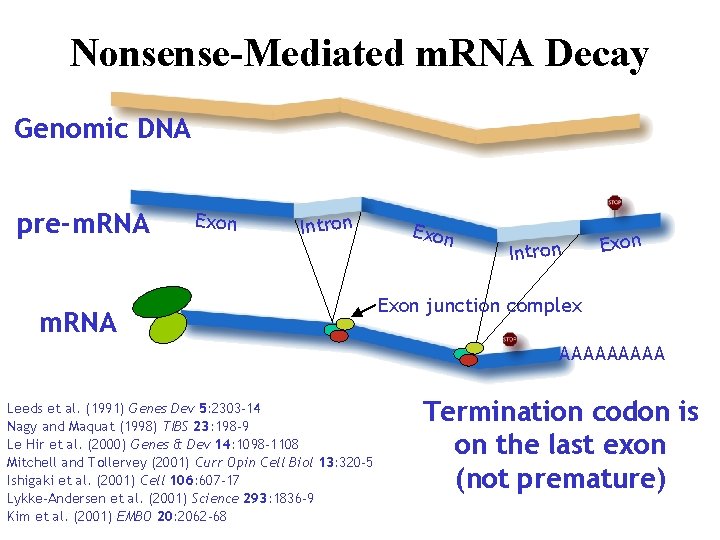
Nonsense-Mediated m. RNA Decay Genomic DNA pre-m. RNA Intron Gm m. RNA Exon Intron Exon junction complex AAAAA Leeds et al. (1991) Genes Dev 5: 2303 -14 Nagy and Maquat (1998) TIBS 23: 198 -9 Le Hir et al. (2000) Genes & Dev 14: 1098 -1108 Mitchell and Tollervey (2001) Curr Opin Cell Biol 13: 320 -5 Ishigaki et al. (2001) Cell 106: 607 -17 Lykke-Andersen et al. (2001) Science 293: 1836 -9 Kim et al. (2001) EMBO 20: 2062 -68 Termination codon is on the last exon (not premature)

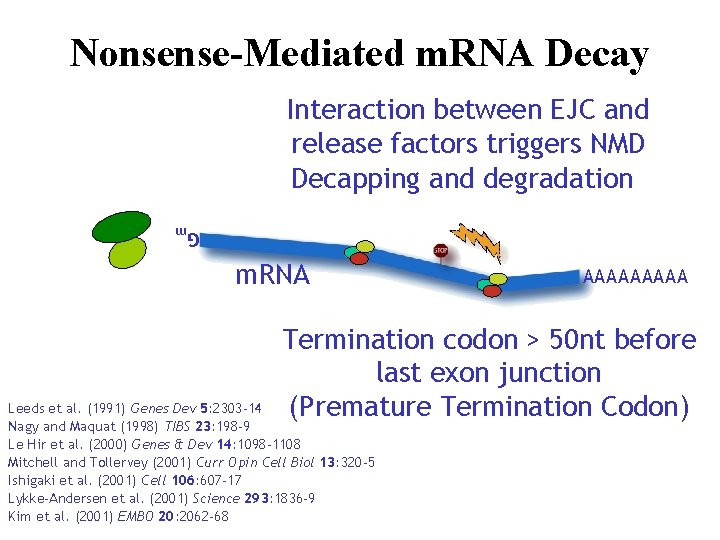
Nonsense-Mediated m. RNA Decay Interaction between EJC and release factors triggers NMD Decapping and degradation Gm m. RNA AAAAA Termination codon > 50 nt before last exon junction (Premature Termination Codon) Leeds et al. (1991) Genes Dev 5: 2303 -14 Nagy and Maquat (1998) TIBS 23: 198 -9 Le Hir et al. (2000) Genes & Dev 14: 1098 -1108 Mitchell and Tollervey (2001) Curr Opin Cell Biol 13: 320 -5 Ishigaki et al. (2001) Cell 106: 607 -17 Lykke-Andersen et al. (2001) Science 293: 1836 -9 Kim et al. (2001) EMBO 20: 2062 -68

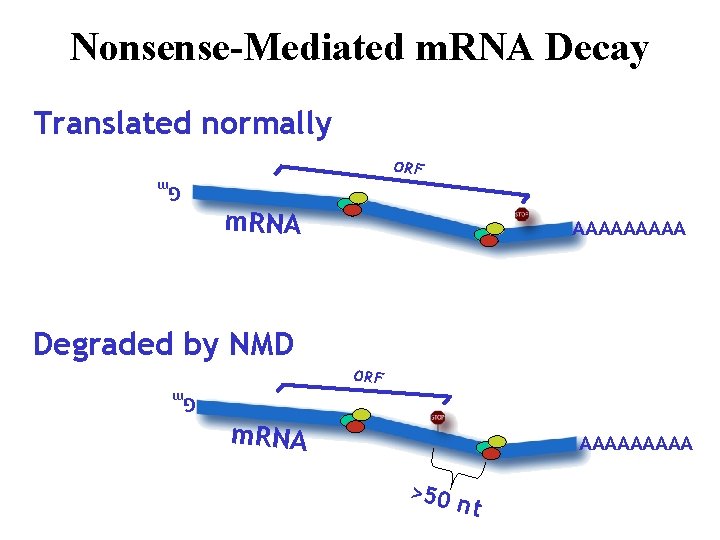
Nonsense-Mediated m. RNA Decay Translated normally ORF m. RNA AAAAA Gm Degraded by NMD ORF m. RNA AAAAA >50 nt Gm

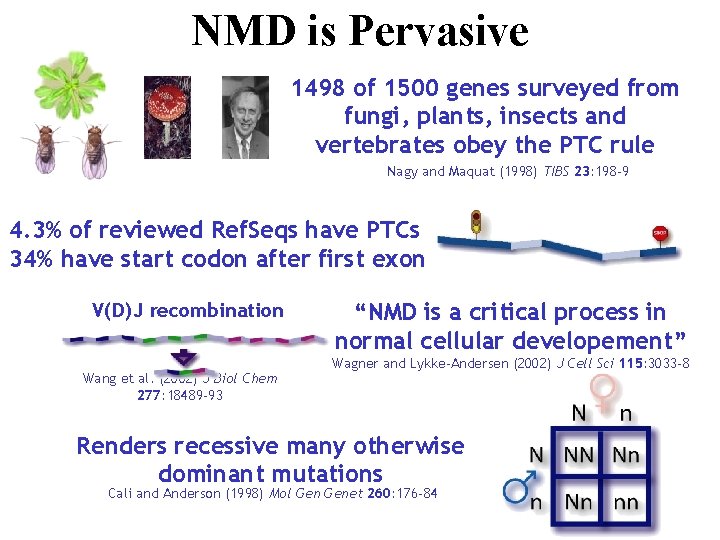
NMD is Pervasive 1498 of 1500 genes surveyed from fungi, plants, insects and vertebrates obey the PTC rule Nagy and Maquat (1998) TIBS 23: 198 -9 4. 3% of reviewed Ref. Seqs have PTCs 34% have start codon after first exon V(D)J recombination Wang et al. (2002) J Biol Chem 277: 18489 -93 “NMD is a critical process in normal cellular developement” Wagner and Lykke-Andersen (2002) J Cell Sci 115: 3033 -8 Renders recessive many otherwise dominant mutations Cali and Anderson (1998) Mol Genet 260: 176 -84

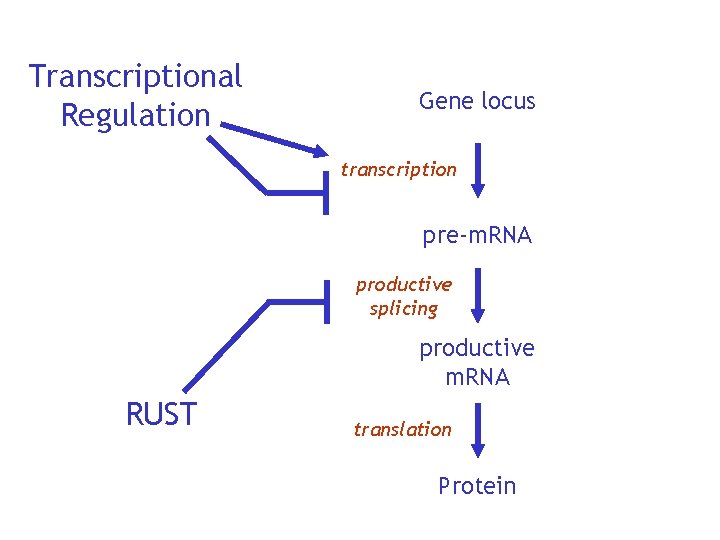
Transcriptional Regulation Gene locus transcription pre-m. RNA productive splicing productive m. RNA RUST translation Protein

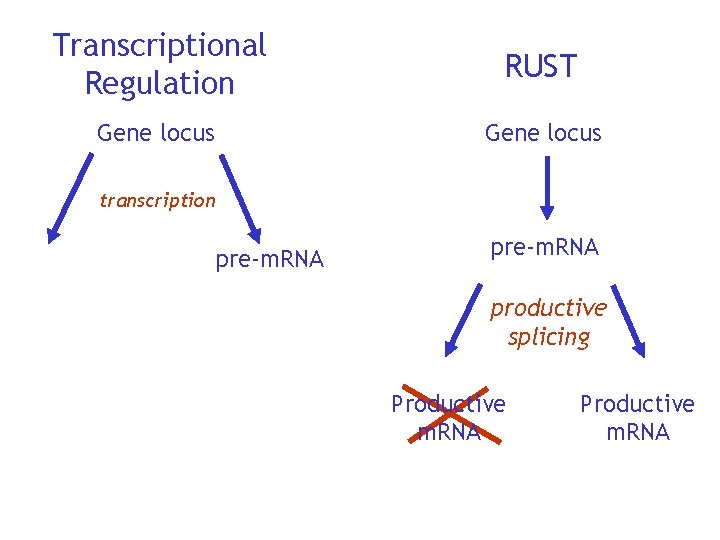
Transcriptional Regulation RUST Gene locus transcription pre-m. RNA productive splicing Productive m. RNA

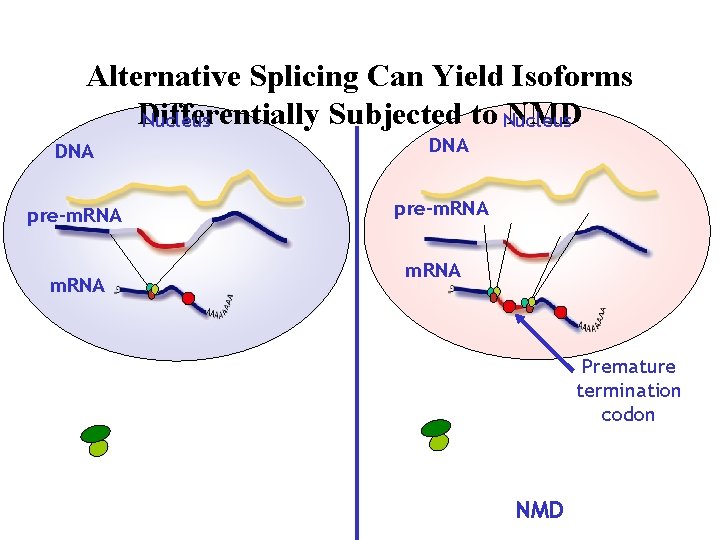
Alternative Splicing Can Yield Isoforms Differentially Subjected to Nucleus NMD Nucleus DNA pre-m. RNA Premature termination codon NMD

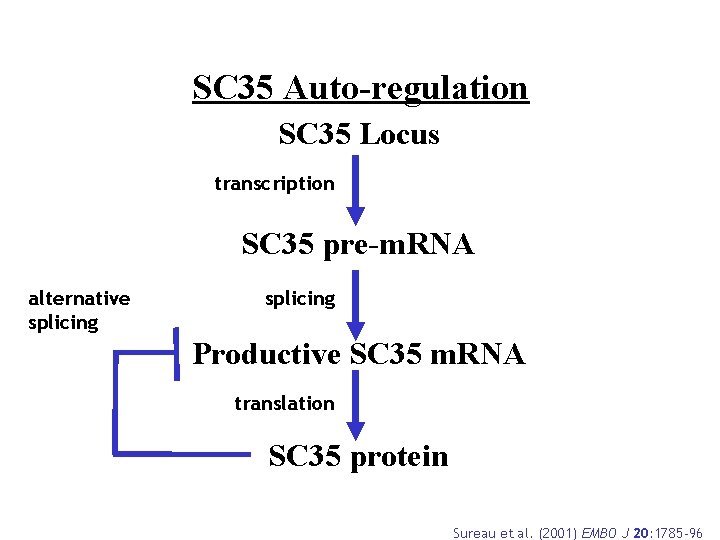
SC 35 Auto-regulation SC 35 Locus transcription SC 35 pre-m. RNA alternative splicing Productive SC 35 m. RNA translation SC 35 protein Sureau et al. (2001) EMBO J 20: 1785 -96

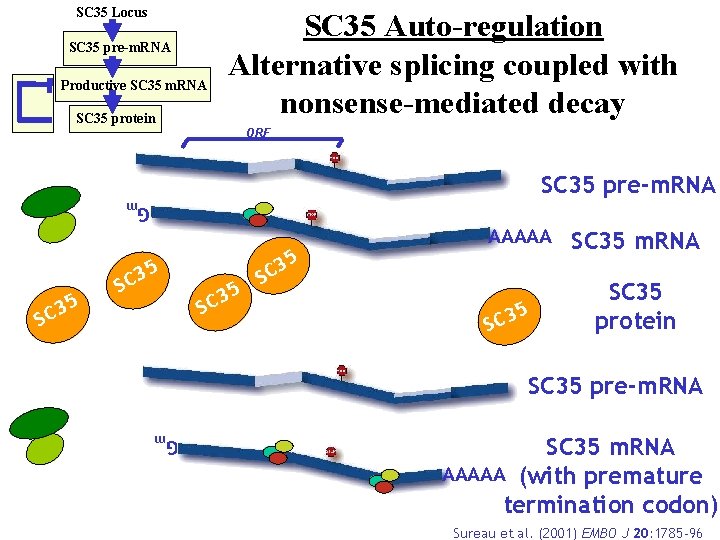
SC 35 Locus SC 35 pre-m. RNA Productive SC 35 m. RNA SC 35 protein SC 35 Auto-regulation Alternative splicing coupled with nonsense-mediated decay ORF SC 35 pre-m. RNA Gm 35 5 3 SC 5 SC 35 m. RNA 5 SC 35 protein SC 35 pre-m. RNA Gm SC 35 SC 3 SC AAAAA SC 35 m. RNA AAAAA (with premature termination codon) Sureau et al. (2001) EMBO J 20: 1785 -96

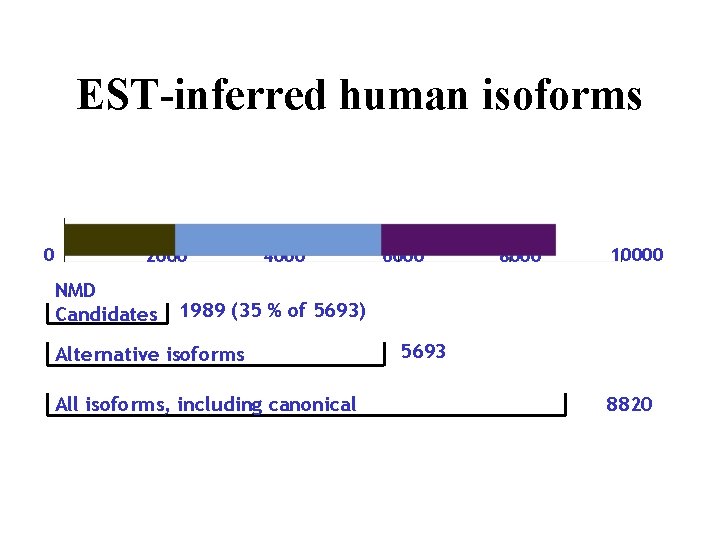
EST-inferred human isoforms 0 2000 NMD Candidates 4000 6000 8000 10000 1989 (35 % of 5693) Alternative isoforms All isoforms, including canonical 5693 8820

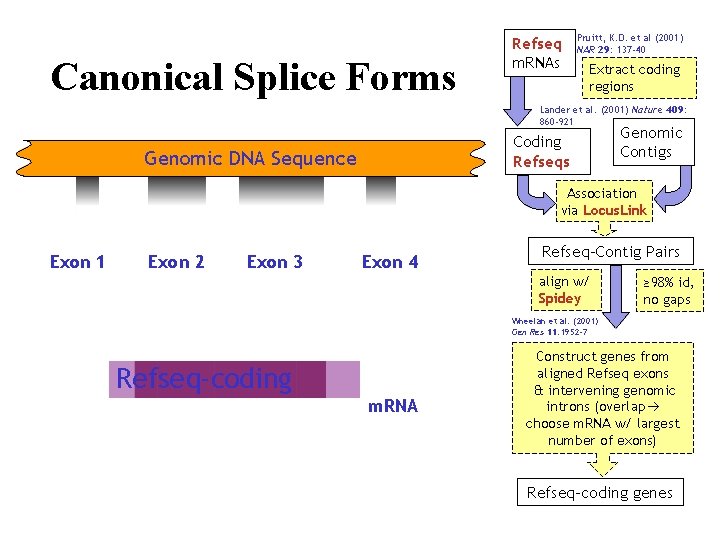
Canonical Splice Forms Refseq m. RNAs Pruitt, K. D. et al (2001) NAR 29: 137 -40 Extract coding regions Lander et al. (2001) Nature 409: 860 -921 Coding Refseqs Genomic DNA Sequence Genomic Contigs Association via Locus. Link Exon 1 Exon 2 Exon 3 Exon 4 Refseq-Contig Pairs align w/ Spidey ≥ 98% id, no gaps Wheelan et al. (2001) Gen Res 11: 1952 -7 Refseq-coding gene m. RNA Construct genes from aligned Refseq exons & intervening genomic introns (overlap choose m. RNA w/ largest number of exons) Refseq-coding genes

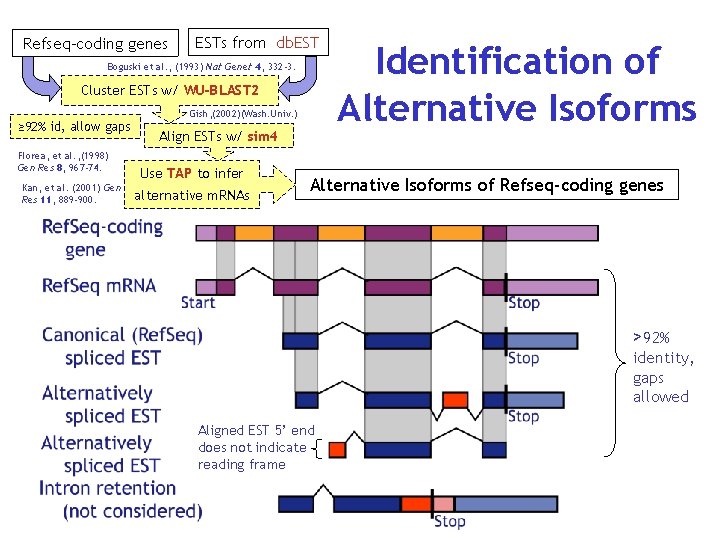
Refseq-coding genes ESTs from db. EST Boguski et al. , (1993) Nat Genet 4, 332 -3. Cluster ESTs w/ WU-BLAST 2 ≥ 92% id, allow gaps Florea, et al. , (1998) Gen Res 8, 967 -74. Kan, et al. (2001) Gen Res 11, 889 -900. Gish, (2002)(Wash. Univ. ) Align ESTs w/ sim 4 Use TAP to infer alternative m. RNAs Identification of Alternative Isoforms of Refseq-coding genes >92% identity, gaps allowed Aligned EST 5’ end does not indicate reading frame

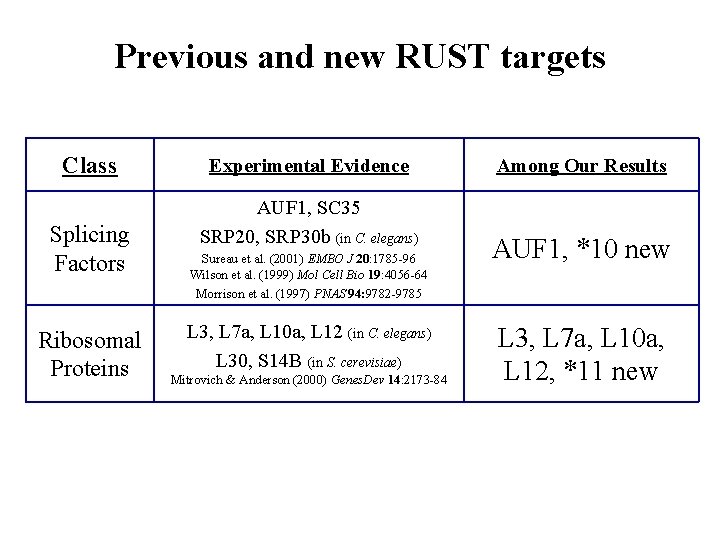
Previous and new RUST targets Class Splicing Factors Ribosomal Proteins Experimental Evidence AUF 1, SC 35 SRP 20, SRP 30 b (in C. elegans) Sureau et al. (2001) EMBO J 20: 1785 -96 Wilson et al. (1999) Mol Cell Bio 19: 4056 -64 Morrison et al. (1997) PNAS 94: 9782 -9785 L 3, L 7 a, L 10 a, L 12 (in C. elegans) L 30, S 14 B (in S. cerevisiae) Mitrovich & Anderson (2000) Genes. Dev 14: 2173 -84 Among Our Results AUF 1, *10 new L 3, L 7 a, L 10 a, L 12, *11 new

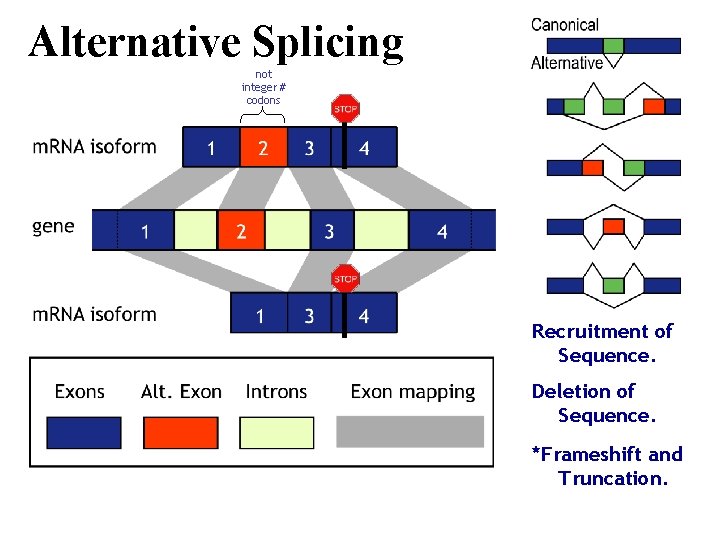
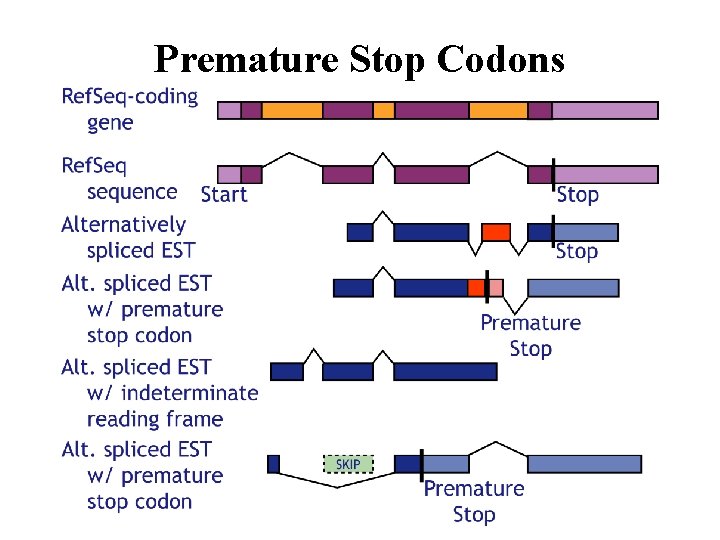
Alternative Splicing not integer # codons Recruitment of Sequence. Deletion of Sequence. *Frameshift and Truncation.

Premature Stop Codons

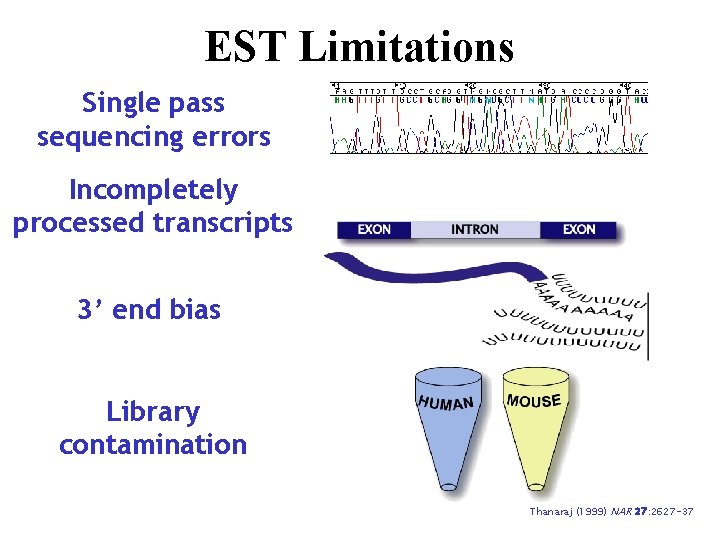
EST Limitations Single pass sequencing errors Incompletely processed transcripts 3’ end bias Library contamination Thanaraj (1999) NAR 27: 2627 -37

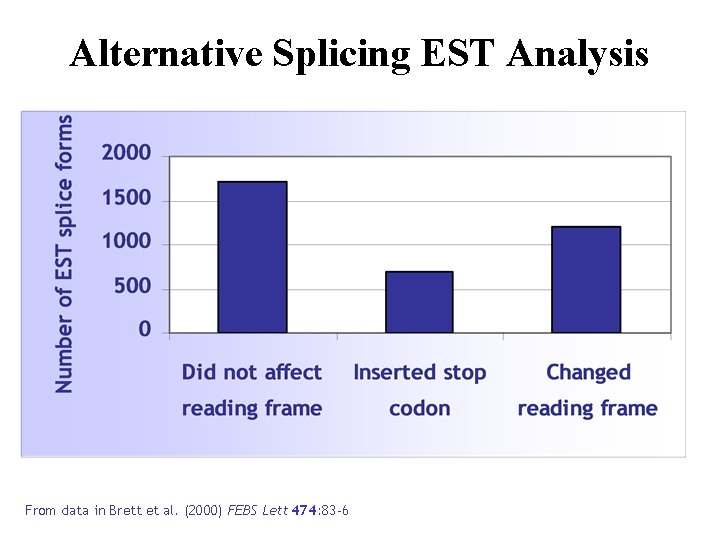
Alternative Splicing EST Analysis From data in Brett et al. (2000) FEBS Lett 474: 83 -6

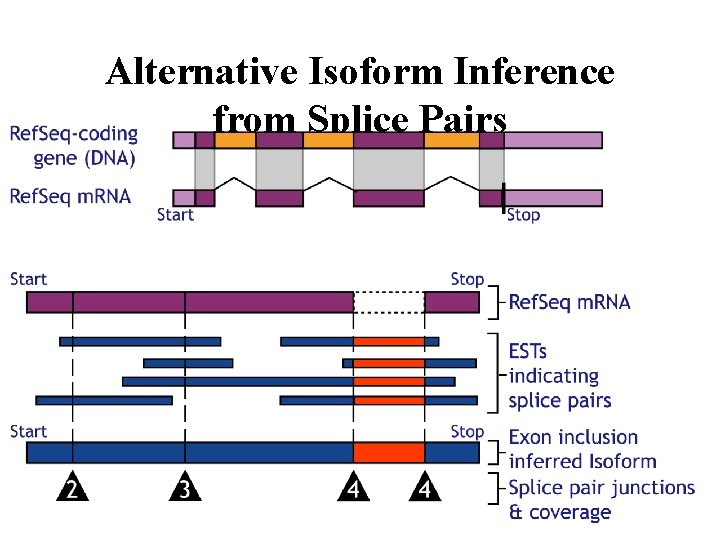
Alternative Isoform Inference from Splice Pairs

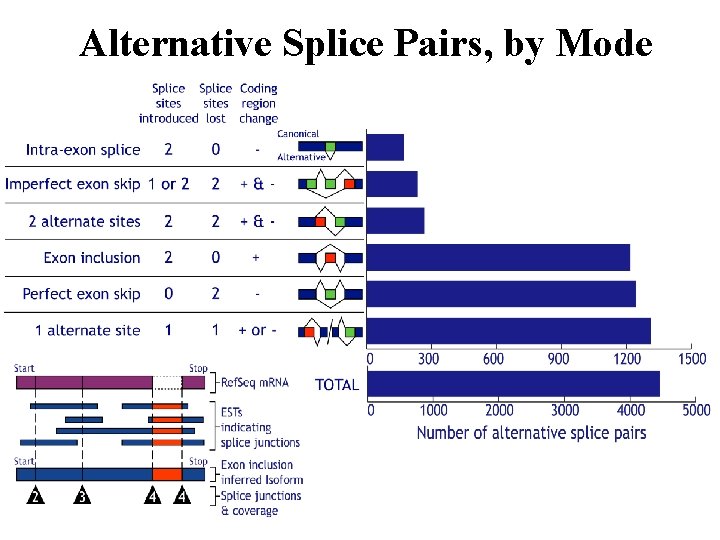
Alternative Splice Pairs, by Mode

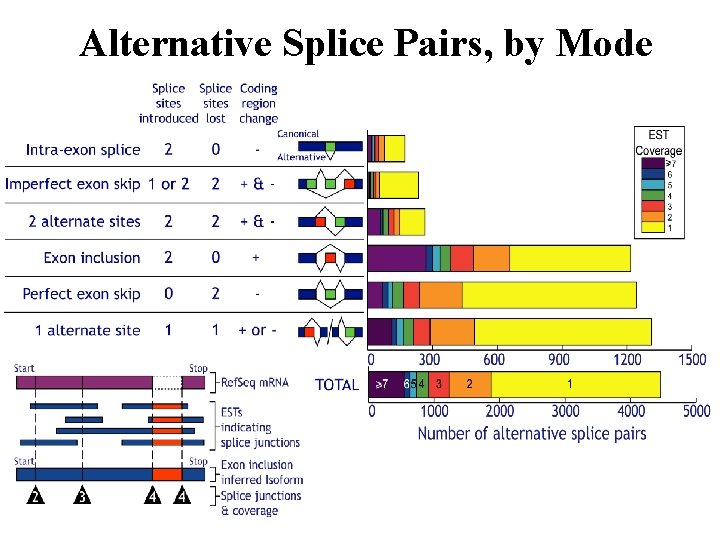
Alternative Splice Pairs, by Mode

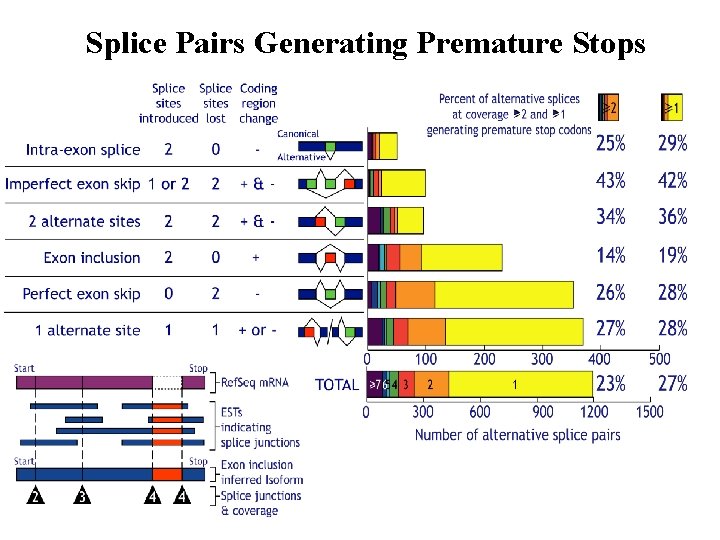
Splice Pairs Generating Premature Stops

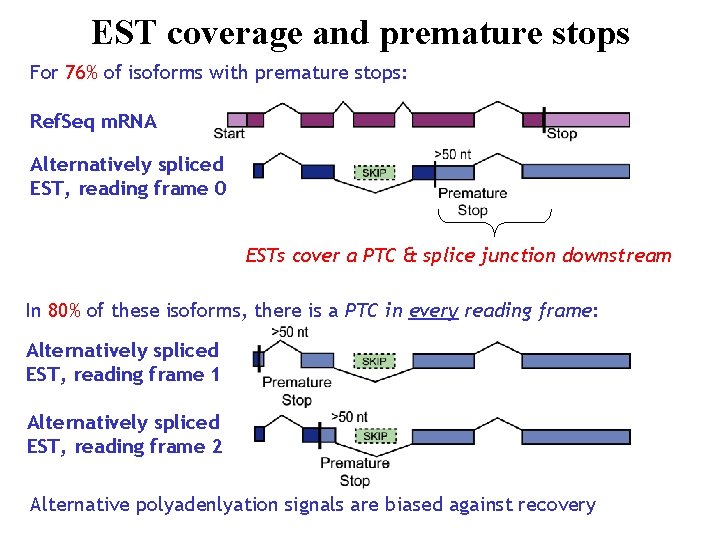
EST coverage and premature stops For 76% of isoforms with premature stops: Ref. Seq m. RNA Alternatively spliced EST, reading frame 0 ESTs cover a PTC & splice junction downstream In 80% of these isoforms, there is a PTC in every reading frame: Alternatively spliced EST, reading frame 1 Alternatively spliced EST, reading frame 2 Alternative polyadenlyation signals are biased against recovery
