Whats New With NAMD Triumph and Torture with



- Slides: 33

What’s New With NAMD Triumph and Torture with New Platforms Jim Phillips and Chee Wai Lee Theoretical and Computational Biophysics Group http: //www. ks. uiuc. edu/Research/namd/ NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

What is NAMD? • Molecular dynamics and related algorithms – e. g. , minimization, steering, locally enhanced sampling, alchemical and conformational free energy perturbation • Efficient algorithms for full electrostatics • Effective on affordable commodity hardware • Read file formats from standard packages: X-PLOR (NAMD 1. 0), CHARMM (NAMD 2. 0), Amber (NAMD 2. 3), GROMACS (NAMD 2. 4) • Building a complete modeling environment NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

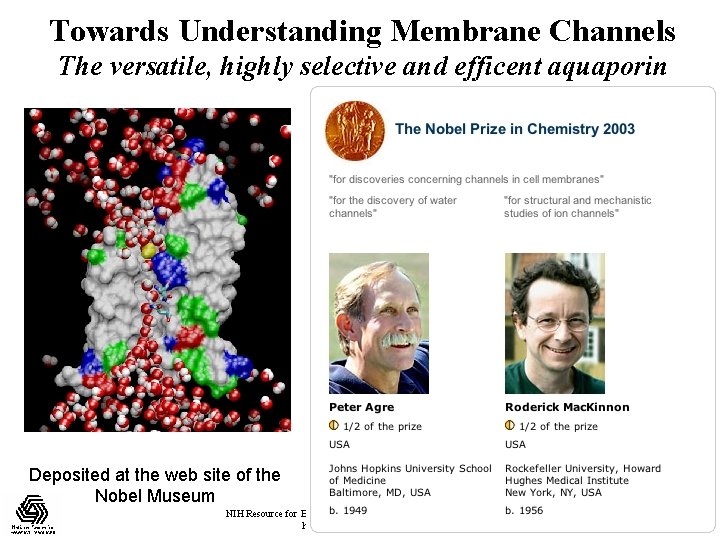
Towards Understanding Membrane Channels The versatile, highly selective and efficent aquaporin Deposited at the web site of the Nobel Museum NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

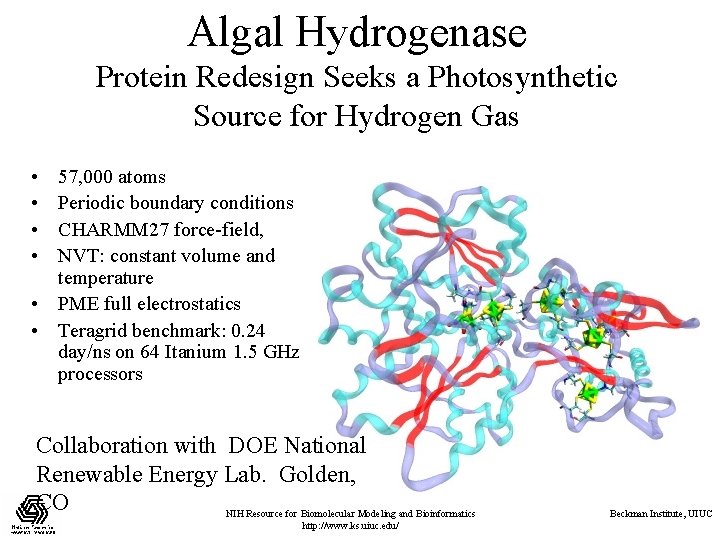
Algal Hydrogenase Protein Redesign Seeks a Photosynthetic Source for Hydrogen Gas • • 57, 000 atoms Periodic boundary conditions CHARMM 27 force-field, NVT: constant volume and temperature • PME full electrostatics • Teragrid benchmark: 0. 24 day/ns on 64 Itanium 1. 5 GHz processors Collaboration with DOE National Renewable Energy Lab. Golden, CO NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

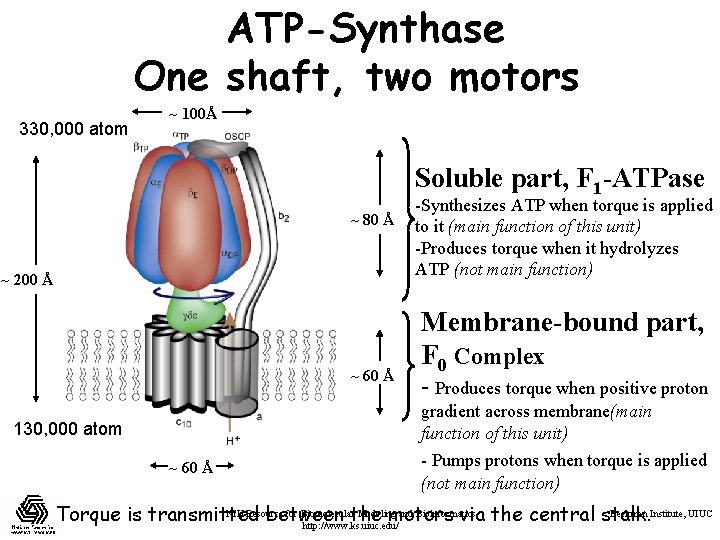
ATP-Synthase One shaft, two motors 330, 000 atom ~ 100Å Soluble part, F 1 -ATPase ~ 80 Å ~ 200 Å ~ 60 Å -Synthesizes ATP when torque is applied to it (main function of this unit) -Produces torque when it hydrolyzes ATP (not main function) Membrane-bound part, F 0 Complex - Produces torque when positive proton gradient across membrane(main function of this unit) 130, 000 atom ~ 60 Å - Pumps protons when torque is applied (not main function) NIH Resource for Biomolecularthe Modeling and Bioinformatics Beckman Institute, UIUC Torque is transmitted between motors via the central stalk. http: //www. ks. uiuc. edu/

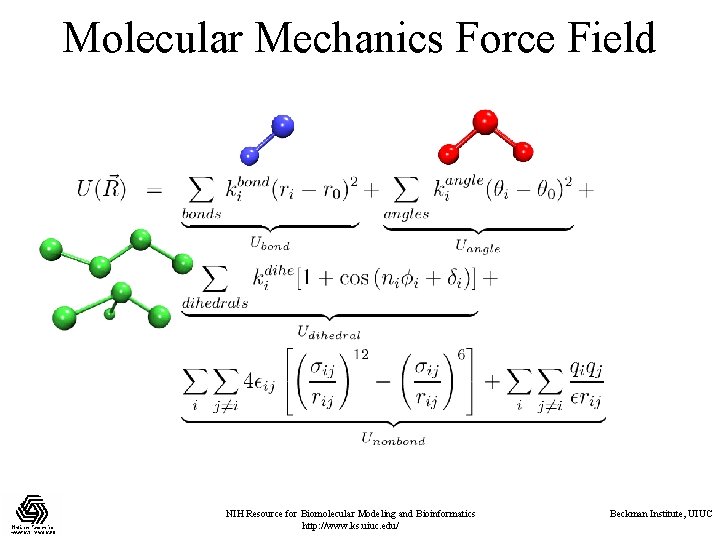
Molecular Mechanics Force Field NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

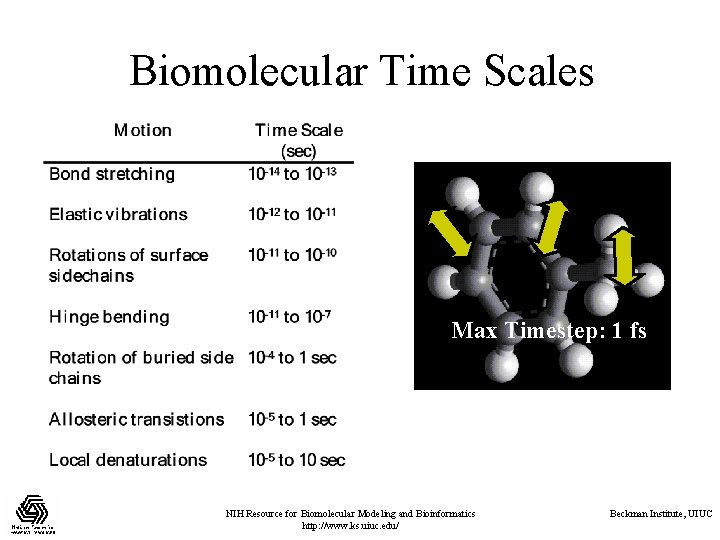
Biomolecular Time Scales Max Timestep: 1 fs NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

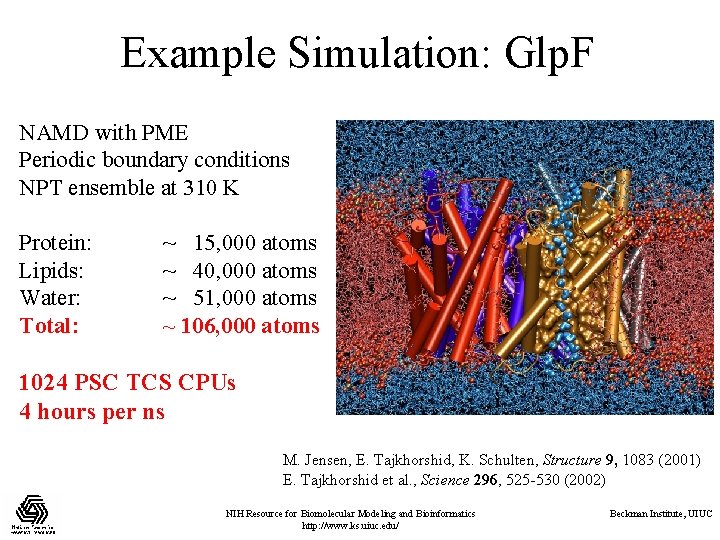
Example Simulation: Glp. F NAMD with PME Periodic boundary conditions NPT ensemble at 310 K Protein: Lipids: Water: Total: ~ 15, 000 atoms ~ 40, 000 atoms ~ 51, 000 atoms ~ 106, 000 atoms 1024 PSC TCS CPUs 4 hours per ns M. Jensen, E. Tajkhorshid, K. Schulten, Structure 9, 1083 (2001) E. Tajkhorshid et al. , Science 296, 525 -530 (2002) NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

Typical Simulation Statistics • • • 100, 000 atoms (including water, lipid) 10 -20 MB of data for entire system 100 A per side periodic cell 12 A cutoff of short-range nonbonded terms 10, 000 timesteps (10 ns) 4 s/step on one processor (1. 3 years total!) NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

Parallel MD: Easy or Hard? • Easy • Hard – – – – – Tiny working data Spatial locality Uniform atom density Persistent repetition Multiple timestepping Sequential timesteps Short iteration time Full electrostatics Fixed problem size NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

Poorly Scaling Approaches • Replicated data – All atom coordinates stored on each processor – Communication/Computation ratio: O(P log P) • Partition the atom array across processors – Nearby atoms may not be on the same processor – C/C ratio: O(P) • Distribute force matrix to processors – Matrix is sparse, non uniform – C/C Ratio: O(sqrt P) NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

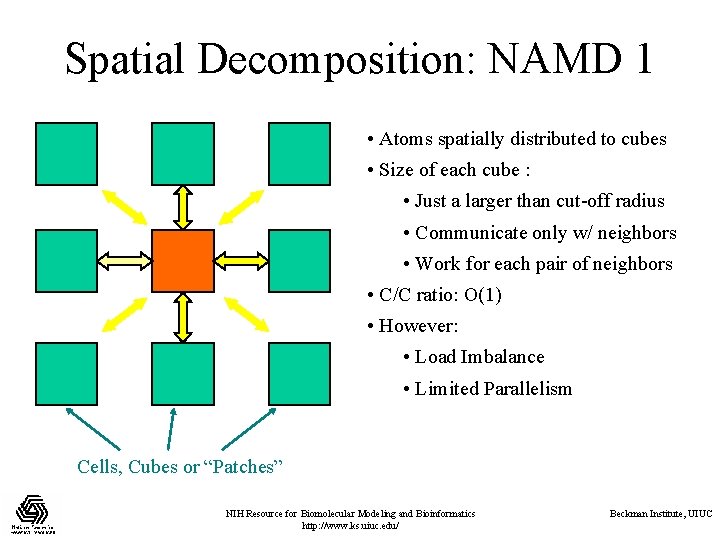
Spatial Decomposition: NAMD 1 • Atoms spatially distributed to cubes • Size of each cube : • Just a larger than cut-off radius • Communicate only w/ neighbors • Work for each pair of neighbors • C/C ratio: O(1) • However: • Load Imbalance • Limited Parallelism Cells, Cubes or “Patches” NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

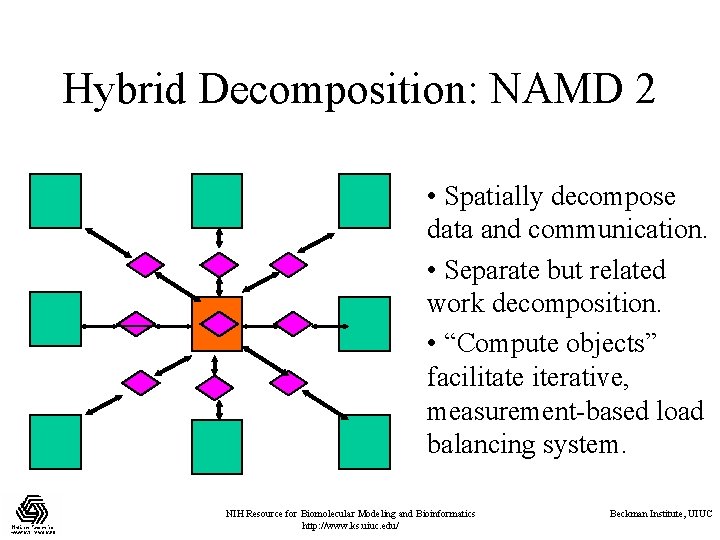
Hybrid Decomposition: NAMD 2 • Spatially decompose data and communication. • Separate but related work decomposition. • “Compute objects” facilitate iterative, measurement-based load balancing system. NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

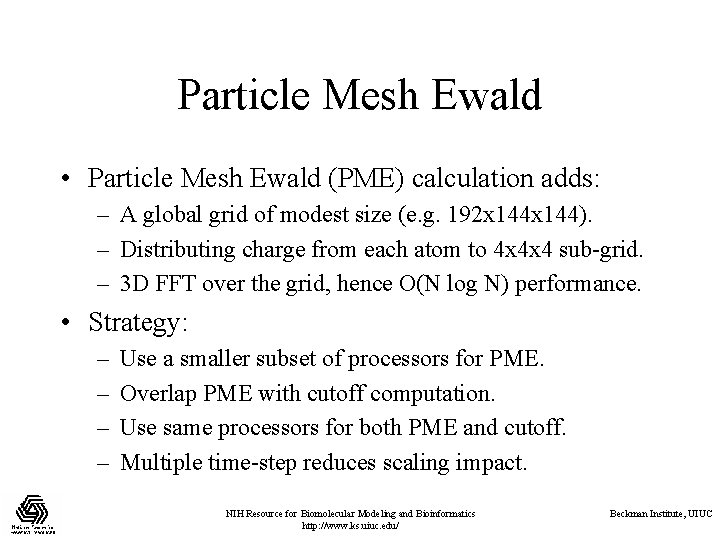
Particle Mesh Ewald • Particle Mesh Ewald (PME) calculation adds: – A global grid of modest size (e. g. 192 x 144). – Distributing charge from each atom to 4 x 4 x 4 sub-grid. – 3 D FFT over the grid, hence O(N log N) performance. • Strategy: – – Use a smaller subset of processors for PME. Overlap PME with cutoff computation. Use same processors for both PME and cutoff. Multiple time-step reduces scaling impact. NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

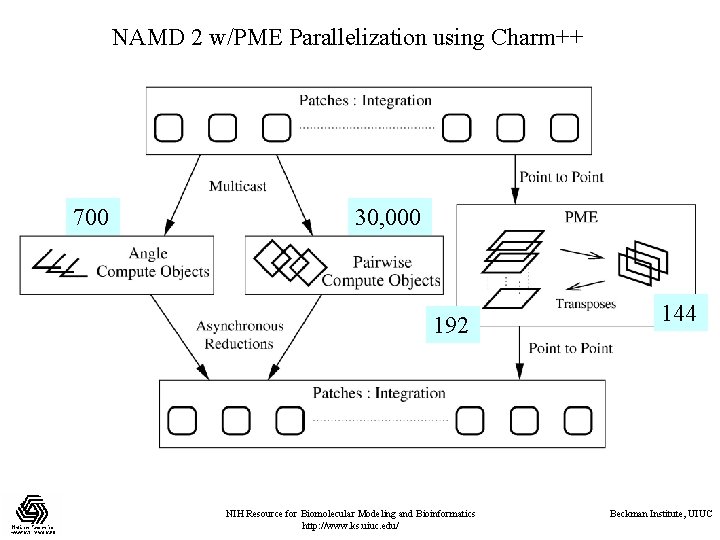
NAMD 2 w/PME Parallelization using Charm++ 700 30, 000 192 NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ 144 Beckman Institute, UIUC

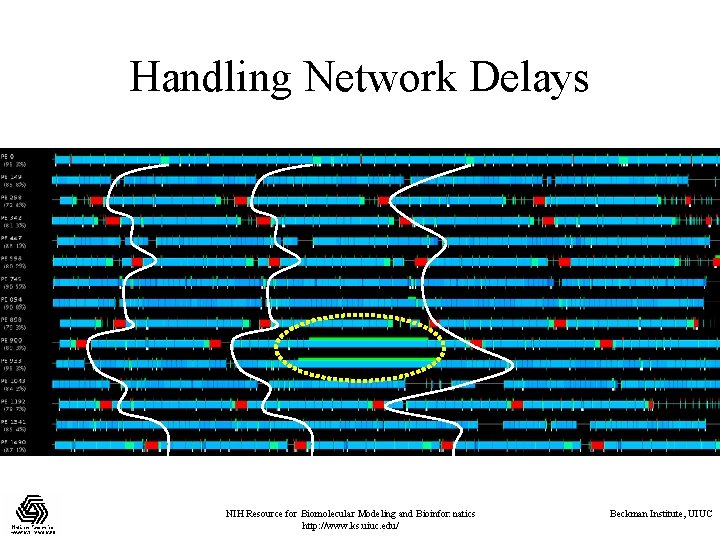
Avoiding Barriers • In NAMD: – The energy reductions were made asynchronous. – No other global barriers are used in cut-off simulations. • This came handy when: – Running on Pittsburgh Lemieux (3000 processors). – The machine (and how Converse uses the network) produced unpredictable, random communication delay. • A send call would remain stuck for 20 ms, for example. – Each timestep, ideally, was 12 -14 ms. NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

Handling Network Delays NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

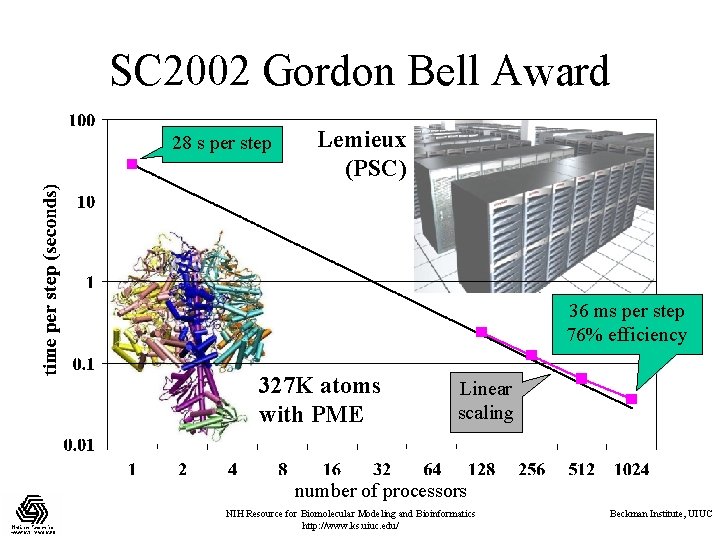
SC 2002 Gordon Bell Award 28 s per step Lemieux (PSC) 36 ms per step 76% efficiency 327 K atoms with PME Linear scaling number of processors NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

Major New Platforms • SGI Altix • Cray XT 3 “Red Storm” • IBM BG/L “Blue Gene” NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

SGI Altix 3000 • • • Itanium-based successor to Origin series 1. 6 GHz Itanium 2 CPUs w/ 9 MB Cache-coherent NUMA shared memory Runs Linux (with some SGI modifications) NCSA has two 512 processor machines NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

Porting NAMD to the Altix • Normal Itanium binary just works. • Best serial performance ever, better than other Itanium platforms (Tera. Grid) at same clock speed. • Building with SGI MPI just works. • setenv MPI_DSM_DISTRIBUTE needed. • Superlinear speedups 16 to 64 processors (good network, running mostly in cache at 64). • Decent scaling to 256 (for Apo. A 1 benchmark). • Intel 8. 1 and later compiler performance issues. NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

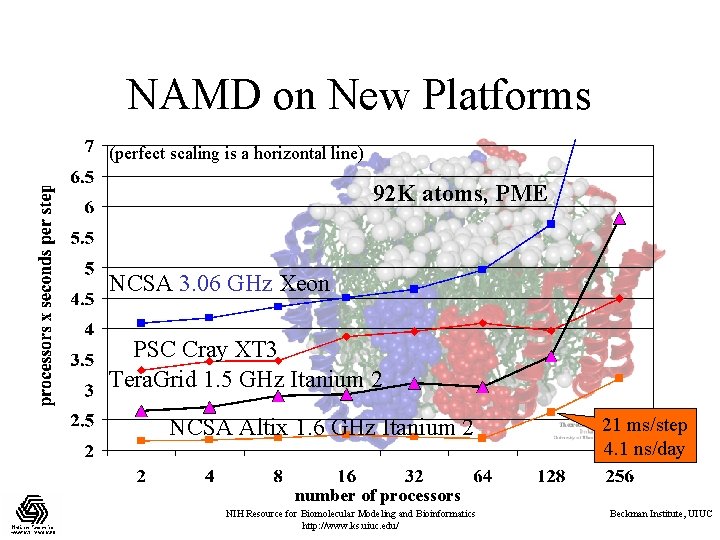
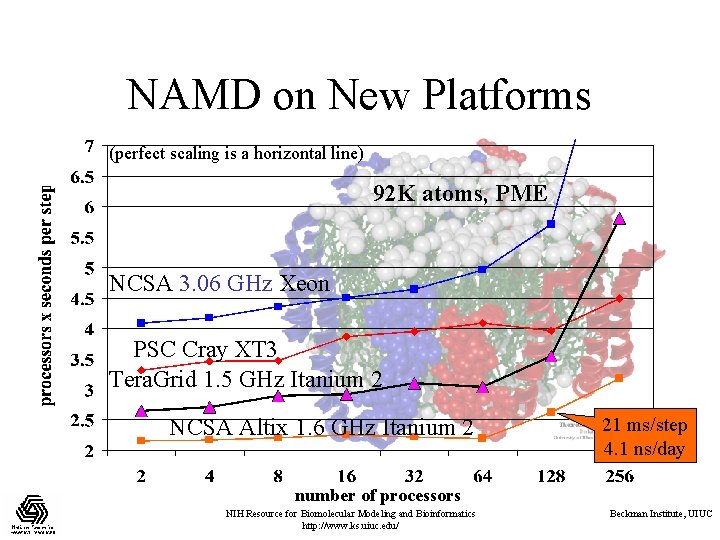
NAMD on New Platforms (perfect scaling is a horizontal line) 92 K atoms, PME NCSA 3. 06 GHz Xeon PSC Cray XT 3 Tera. Grid 1. 5 GHz Itanium 2 NCSA Altix 1. 6 GHz Itanium 2 21 ms/step 4. 1 ns/day number of processors NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

Altix Conclusions • Nice machine, easy to port to – Code must run well on Itanium • Perfect for typical NAMD user – Fastest serial performance – Scales well to typical number of processors – Full environment, no surprises – TCBG’s favorite platform for the past year NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

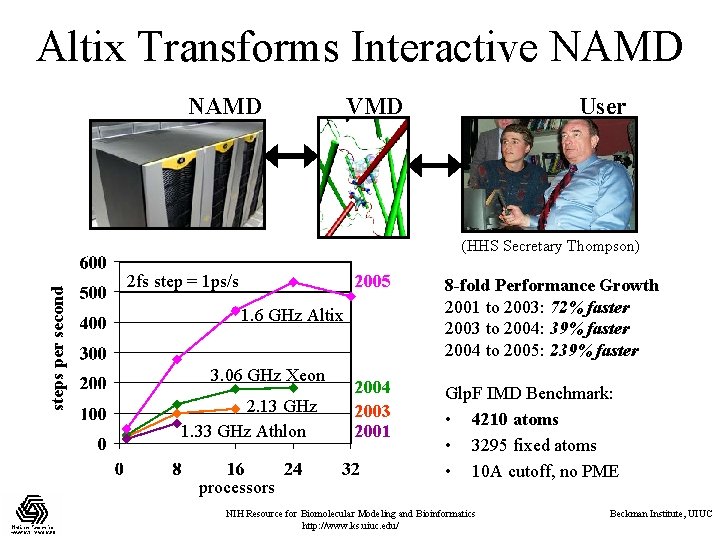
Altix Transforms Interactive NAMD VMD User steps per second (HHS Secretary Thompson) 2 fs step = 1 ps/s 2005 8 -fold Performance Growth 2001 to 2003: 72% faster 2003 to 2004: 39% faster 2004 to 2005: 239% faster 2004 2003 2001 Glp. F IMD Benchmark: • 4210 atoms • 3295 fixed atoms • 10 A cutoff, no PME 1. 6 GHz Altix 3. 06 GHz Xeon 2. 13 GHz 1. 33 GHz Athlon processors NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

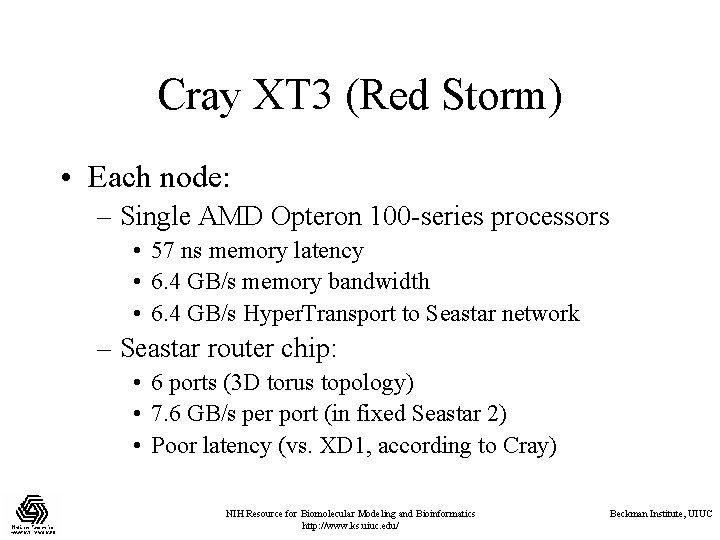
Cray XT 3 (Red Storm) • Each node: – Single AMD Opteron 100 -series processors • 57 ns memory latency • 6. 4 GB/s memory bandwidth • 6. 4 GB/s Hyper. Transport to Seastar network – Seastar router chip: • 6 ports (3 D torus topology) • 7. 6 GB/s per port (in fixed Seastar 2) • Poor latency (vs. XD 1, according to Cray) NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

Cray XT 3 (Red Storm) • • 4 nodes per blade 8 blades per chassis 3 chassis per cabinet, plus one big fan PSC machine (Big Ben) has 22 chassis – 2068 compute processors – Performance boost for TCS system (Lemieux) NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

Cray XT 3 (Red Storm) • Service and I/O nodes run Linux – Normal x 64 -64 binaries just work on them • Compute nodes run Catamount kernel – – – No OS interference for fine-grained parallelism No time sharing…one process at a time No sockets No interrupts No virtual memory System calls forwarded to head node (slow!) NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

Cray XT 3 Porting • Initial compile mostly straightforward – Disable Tcl, sockets, hostname, username code. • Initial runs horribly slow on startup – Almost like memory allocation was O(n 2) – Found docs: • “simplementation of malloc(), optimized for the lightweight kernel and large memory allocations” • Sounds like they assume a stack-based structure • Using –lgmalloc restores sane performance NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

Cray XT 3 Porting • Still somewhat slow on startup – Need to do all I/O to Lustre scratch space – May be better when head node isn’t overloaded • Tried SHMEM port (old T 3 E layer) – New library doesn’t support locks yet – SHMEM was optimized for T 3 E, not XT 3 • Need Tcl for fully functional NAMD – #ifdef out all socket and user info code – Same approach should work on BG/L NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

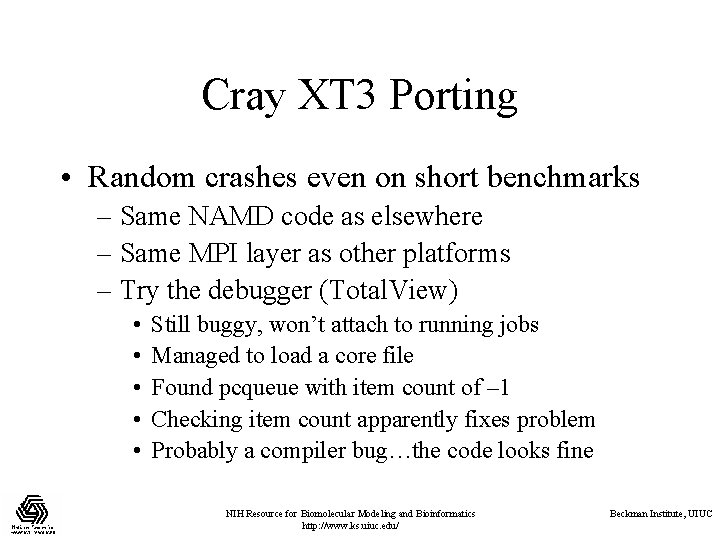
Cray XT 3 Porting • Random crashes even on short benchmarks – Same NAMD code as elsewhere – Same MPI layer as other platforms – Try the debugger (Total. View) • • • Still buggy, won’t attach to running jobs Managed to load a core file Found pcqueue with item count of – 1 Checking item count apparently fixes problem Probably a compiler bug…the code looks fine NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

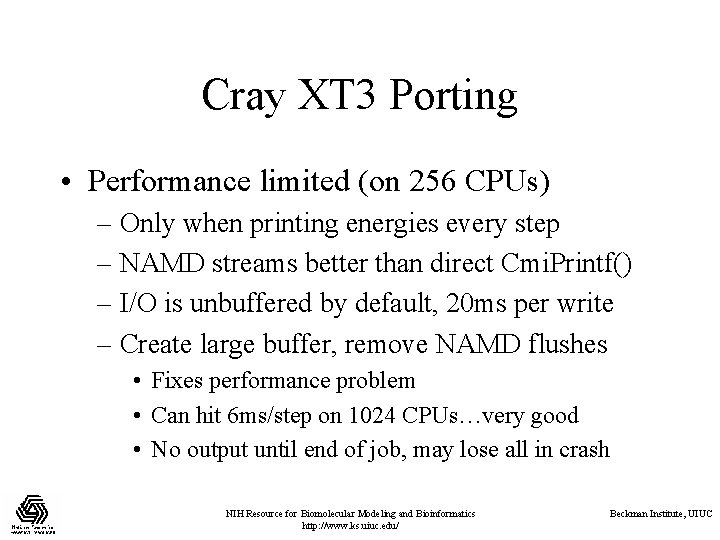
Cray XT 3 Porting • Performance limited (on 256 CPUs) – Only when printing energies every step – NAMD streams better than direct Cmi. Printf() – I/O is unbuffered by default, 20 ms per write – Create large buffer, remove NAMD flushes • Fixes performance problem • Can hit 6 ms/step on 1024 CPUs…very good • No output until end of job, may lose all in crash NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

NAMD on New Platforms (perfect scaling is a horizontal line) 92 K atoms, PME NCSA 3. 06 GHz Xeon PSC Cray XT 3 Tera. Grid 1. 5 GHz Itanium 2 NCSA Altix 1. 6 GHz Itanium 2 21 ms/step 4. 1 ns/day number of processors NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC

Cray XT 3 Conclusions • Serial performance is reasonable – Itanium is faster for NAMD – Opteron requires less tuning work • Scaling is outstanding (eventually) – Low system noise allows 6 ms timesteps – NAMD latency tolerance may help • Lack of OS features annoying, but workable • TCBG’s main allocation for this year NIH Resource for Biomolecular Modeling and Bioinformatics http: //www. ks. uiuc. edu/ Beckman Institute, UIUC
 Namd conference
Namd conference Namd tutorial
Namd tutorial Namd vmd
Namd vmd Triumph pharmaceuticals
Triumph pharmaceuticals Balkandum and balkandee
Balkandum and balkandee The triumph of time analysis
The triumph of time analysis Triumph of bacchus
Triumph of bacchus Chapter 17 section 2 communists triumph in china
Chapter 17 section 2 communists triumph in china The romantic triumph
The romantic triumph Chapter 4 the triumph of industry
Chapter 4 the triumph of industry So gold can easily triumph defeat strongest man
So gold can easily triumph defeat strongest man Triumph group
Triumph group Ktm duke 125 2017 leistungssteigerung
Ktm duke 125 2017 leistungssteigerung Francois boucher triumph of venus
Francois boucher triumph of venus Triumph of death
Triumph of death Triumph house ubc
Triumph house ubc Triumph des willens
Triumph des willens How did triumph
How did triumph Triumph investasi
Triumph investasi Triumph cloud free download
Triumph cloud free download Triumph thruxton mods
Triumph thruxton mods Lesson 3 triumph of parliament in england
Lesson 3 triumph of parliament in england What are carlson's reasons for shooting candy's dog
What are carlson's reasons for shooting candy's dog Dns drip
Dns drip Head crusher device
Head crusher device Iron spider witch torture
Iron spider witch torture Canadian centre for victims of torture
Canadian centre for victims of torture Nnn torture
Nnn torture Why did waverly ask her mother about chinese torture
Why did waverly ask her mother about chinese torture Knee crushing torture
Knee crushing torture Torture
Torture Squassation torture
Squassation torture Tammy homolka
Tammy homolka Romeo and juliet quote test
Romeo and juliet quote test