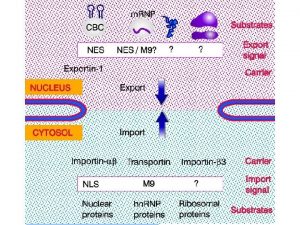
Transport through the nuclear pores The NLS and



















- Slides: 26



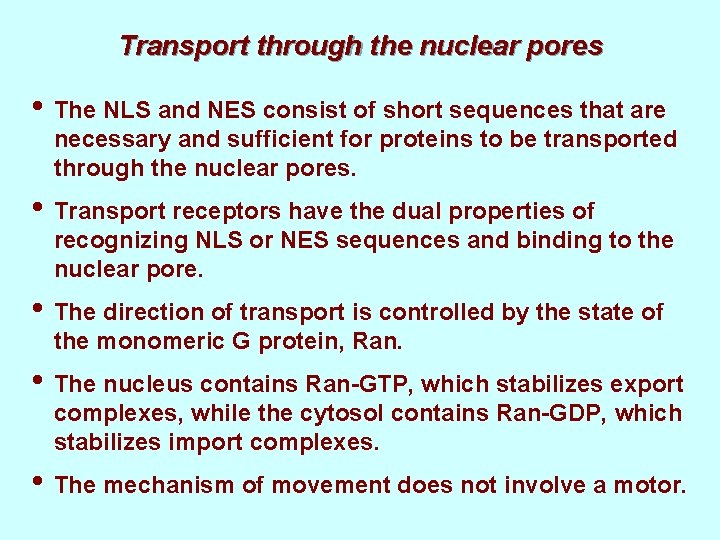
Transport through the nuclear pores • The NLS and NES consist of short sequences that are necessary and sufficient for proteins to be transported through the nuclear pores. • Transport receptors have the dual properties of recognizing NLS or NES sequences and binding to the nuclear pore. • The direction of transport is controlled by the state of the monomeric G protein, Ran. • The nucleus contains Ran-GTP, which stabilizes export complexes, while the cytosol contains Ran-GDP, which stabilizes import complexes. • The mechanism of movement does not involve a motor.


Biologia molecolare - Robert F. Weaver Copyright © 2005 – The Mc. Graw-Hill Companies srl



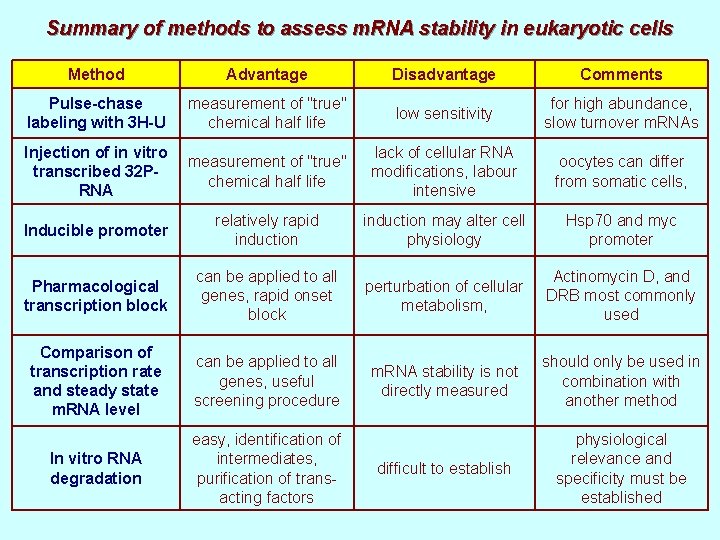
Summary of methods to assess m. RNA stability in eukaryotic cells Method Advantage Disadvantage Comments Pulse-chase labeling with 3 H-U measurement of "true" chemical half life low sensitivity for high abundance, slow turnover m. RNAs Injection of in vitro transcribed 32 PRNA measurement of "true" chemical half life lack of cellular RNA modifications, labour intensive oocytes can differ from somatic cells, Inducible promoter relatively rapid induction may alter cell physiology Hsp 70 and myc promoter Pharmacological transcription block can be applied to all genes, rapid onset block perturbation of cellular metabolism, Actinomycin D, and DRB most commonly used Comparison of transcription rate and steady state m. RNA level can be applied to all genes, useful screening procedure m. RNA stability is not directly measured should only be used in combination with another method In vitro RNA degradation easy, identification of intermediates, purification of transacting factors difficult to establish physiological relevance and specificity must be established


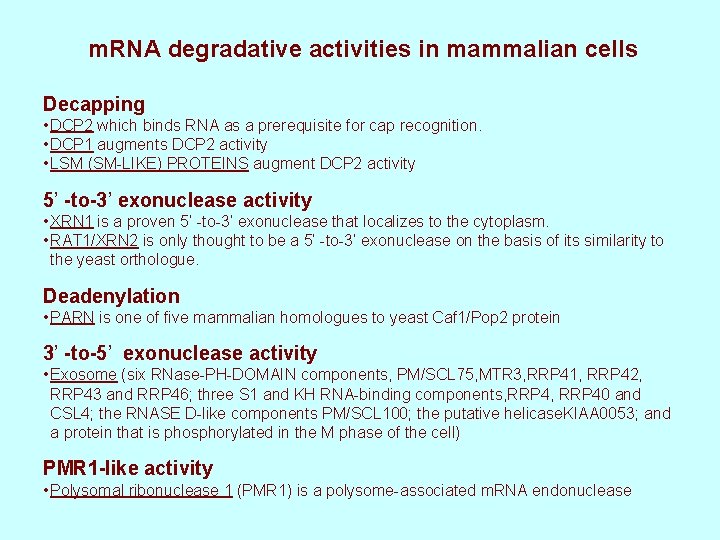
m. RNA degradative activities in mammalian cells Decapping • DCP 2 which binds RNA as a prerequisite for cap recognition. • DCP 1 augments DCP 2 activity • LSM (SM-LIKE) PROTEINS augment DCP 2 activity 5’ -to-3’ exonuclease activity • XRN 1 is a proven 5’ -to-3’ exonuclease that localizes to the cytoplasm. • RAT 1/XRN 2 is only thought to be a 5’ -to-3’ exonuclease on the basis of its similarity to the yeast orthologue. Deadenylation • PARN is one of five mammalian homologues to yeast Caf 1/Pop 2 protein 3’ -to-5’ exonuclease activity • Exosome (six RNase-PH-DOMAIN components, PM/SCL 75, MTR 3, RRP 41, RRP 42, RRP 43 and RRP 46; three S 1 and KH RNA-binding components, RRP 40 and CSL 4; the RNASE D-like components PM/SCL 100; the putative helicase. KIAA 0053; and a protein that is phosphorylated in the M phase of the cell) PMR 1 -like activity • Polysomal ribonuclease 1 (PMR 1) is a polysome-associated m. RNA endonuclease



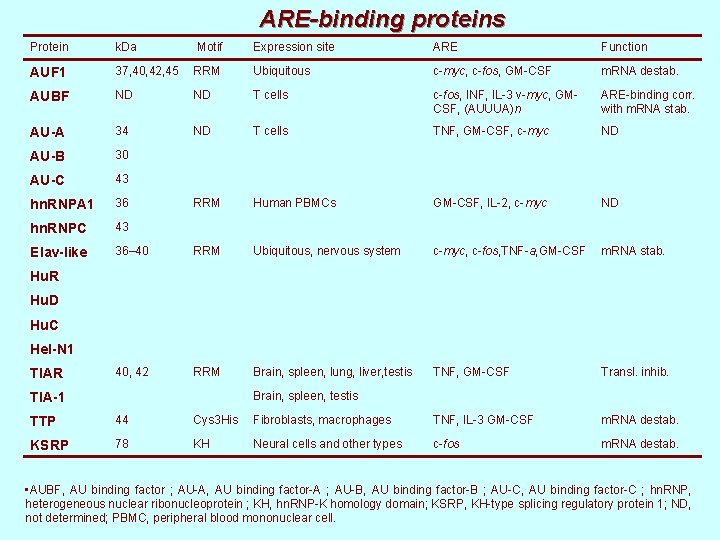
ARE-binding proteins Protein k. Da Motif Expression site ARE Function AUF 1 37, 40, 42, 45 RRM Ubiquitous c-myc, c-fos, GM-CSF m. RNA destab. AUBF ND ND T cells c-fos, INF, IL-3 v-myc, GMCSF, (AUUUA)n ARE-binding corr. with m. RNA stab. AU-A 34 ND T cells TNF, GM-CSF, c-myc ND AU-B 30 AU-C 43 hn. RNPA 1 36 RRM Human PBMCs GM-CSF, IL-2, c-myc ND hn. RNPC 43 Elav-like 36– 40 RRM Ubiquitous, nervous system c-myc, c-fos, TNF-a, GM-CSF m. RNA stab. 40, 42 RRM Brain, spleen, lung, liver, testis TNF, GM-CSF Transl. inhib. Hu. R Hu. D Hu. C Hel-N 1 TIAR Brain, spleen, testis TIA-1 TTP 44 Cys 3 His Fibroblasts, macrophages TNF, IL-3 GM-CSF m. RNA destab. KSRP 78 KH Neural cells and other types c-fos m. RNA destab. • AUBF, AU binding factor ; AU-A, AU binding factor-A ; AU-B, AU binding factor-B ; AU-C, AU binding factor-C ; hn. RNP, heterogeneous nuclear ribonucleoprotein ; KH, hn. RNP-K homology domain; KSRP, KH-type splicing regulatory protein 1; ND, not determined; PBMC, peripheral blood mononuclear cell.










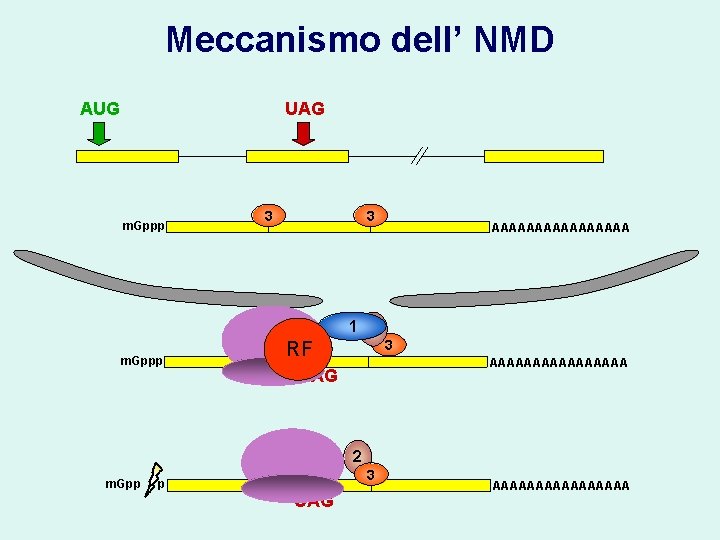
Meccanismo dell’ NMD AUG UAG m. Gppp 3 22 m. Gppp 3 3 3 AAAAAAAA 1 2 RF 3 AAAAAAAA UAG 2 m. Gpp p 3 UAG AAAAAAAA

RNA interference • Meccanismo • Significato biologico • Strumento di analisi della funzione dei geni

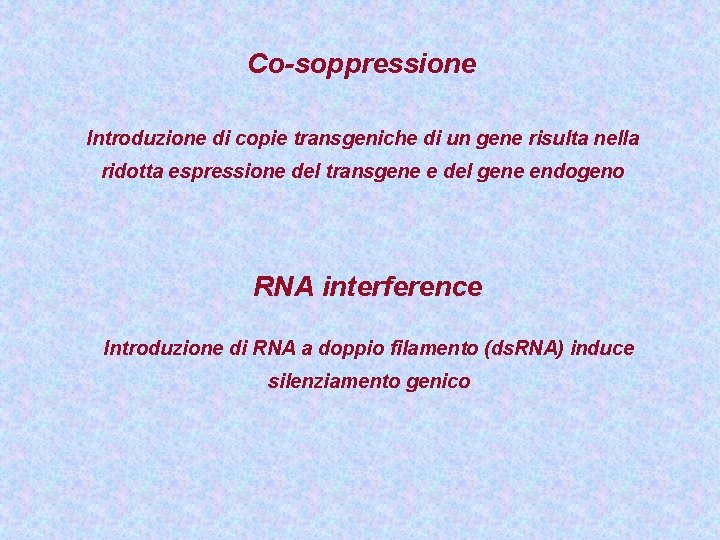
Co-soppressione Introduzione di copie transgeniche di un gene risulta nella ridotta espressione del transgene e del gene endogeno RNA interference Introduzione di RNA a doppio filamento (ds. RNA) induce silenziamento genico

Componenti dell’RNAi • si. RNA (small interfering RNA) = frammenti di RNA 21 -25 nt • Dicer = endoribonucleasi specifica per ds. RNA (tipo RNasi III) • RISC (RNA-induced silencing complex) = complesso ribonucleoproteico • Rd. RP (RNA-dependent RNA polymerase)
 Nucleus diagram
Nucleus diagram Nuclear pores
Nuclear pores Organelle that modifies sorts and packages proteins
Organelle that modifies sorts and packages proteins Nls metatron
Nls metatron Sequenze segnale
Sequenze segnale Oil pores
Oil pores Teana expert boost invisible pores
Teana expert boost invisible pores Lesson 15 nuclear quest nuclear reactions
Lesson 15 nuclear quest nuclear reactions Fisión nuclear vs fision nuclear
Fisión nuclear vs fision nuclear World nuclear transport institute
World nuclear transport institute Symport antiport uniport
Symport antiport uniport Primary active transport and secondary active transport
Primary active transport and secondary active transport What is passive transport
What is passive transport Conversion in timber
Conversion in timber Passive transport vs active transport venn diagram
Passive transport vs active transport venn diagram Active vs passive transport venn diagram
Active vs passive transport venn diagram Endocytosis vs exocytosis
Endocytosis vs exocytosis Primary active transport vs secondary active transport
Primary active transport vs secondary active transport Bioflix activity membrane transport active transport
Bioflix activity membrane transport active transport Bioflix activity membrane transport diffusion
Bioflix activity membrane transport diffusion Through one man
Through one man Through and through furcation
Through and through furcation Poem night of the scorpion
Poem night of the scorpion Fission and fusion similarities
Fission and fusion similarities Quantum and nuclear physics
Quantum and nuclear physics Pron and cons
Pron and cons Difference between nuclear family and joint family
Difference between nuclear family and joint family