Transcriptomics Data Visualization Using Partek Flow Software MAR






- Slides: 30

Transcriptomics Data Visualization Using Partek Flow Software MAR 27, 2019 ANSUMAN CHATTOPADHYAY, PHD ASST DIRECTOR, MOLECULAR BIOLOGY INFORMATION SERVICE HEALTH SCIENCES LIBRARY SYSTEM UNIVERSITY OF PITTSBURGH ANSUMAN@PITT. EDU SRI CHAPARALA, MS BIOINFORMATICS SPECIALIST HEALTH SCIENCES LIBRARY SYSTEM UNIVERSITY OF PITTSBURGH CHAPA 28@PITT. EDU

Workshop Page http: //hsls. libguides. com/molbioworkshops/bulkrnaseqclc

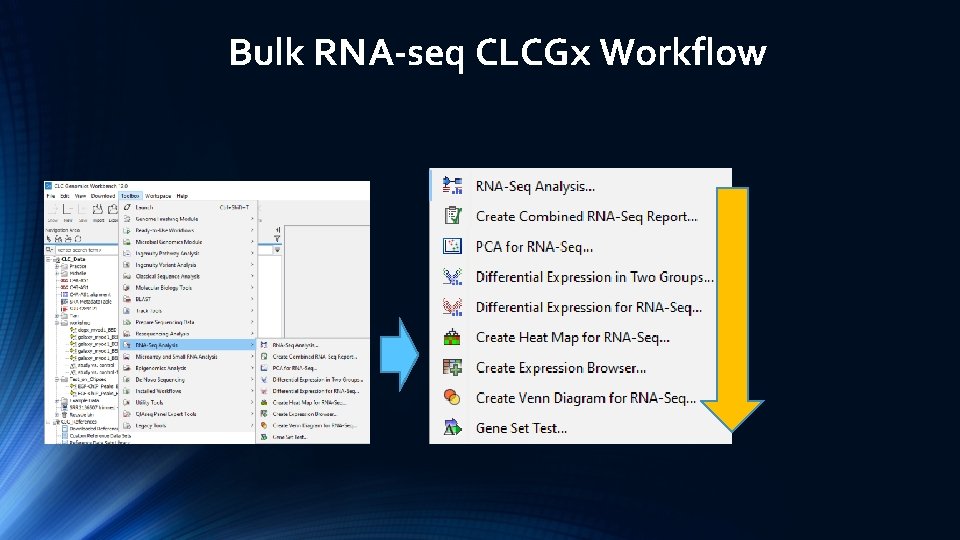
Bulk RNA-seq CLCGx Workflow

Bulk RNA-seq workshop topics • Brief introduction to RNA-Seq experiments • Analyze RNA-seq data • Dexamethasone treatment on airway smooth muscle cells (Himes et al. PLos One 2014) • Download seq reads from EBI-ENA/NCBI SRA • Import reads to CLC Genomics Workbench • Align reads to Reference Genome • Estimate expressions in the gene level • Estimate expressions in the transcript isoform level • Statistical analysis of the differential expressed genes and transcripts • Create Heat Map, Volcano Plots, and Venn Diagram

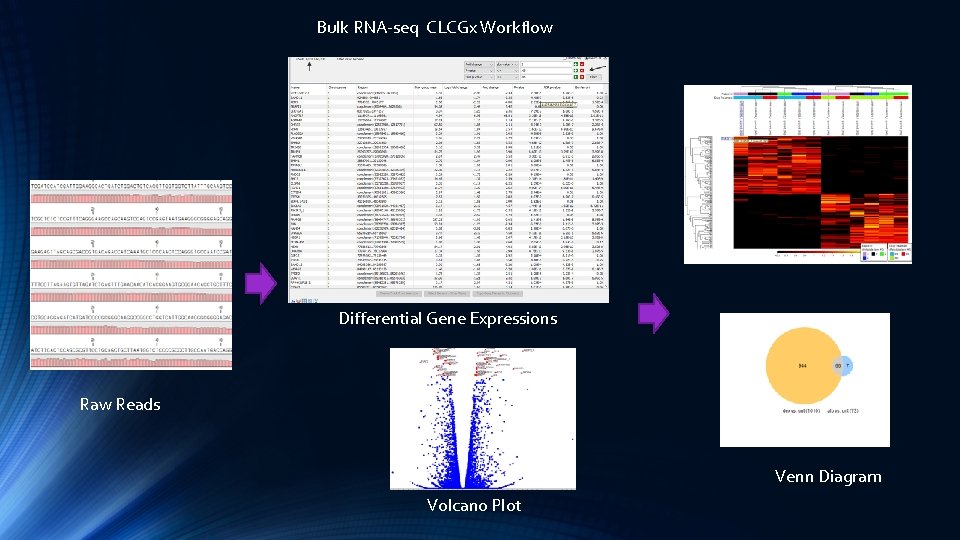
Bulk RNA-seq CLCGx Workflow Differential Gene Expressions Raw Reads Venn Diagram Volcano Plot

Partek Flow software http: //www. partek. com/partek-flow/

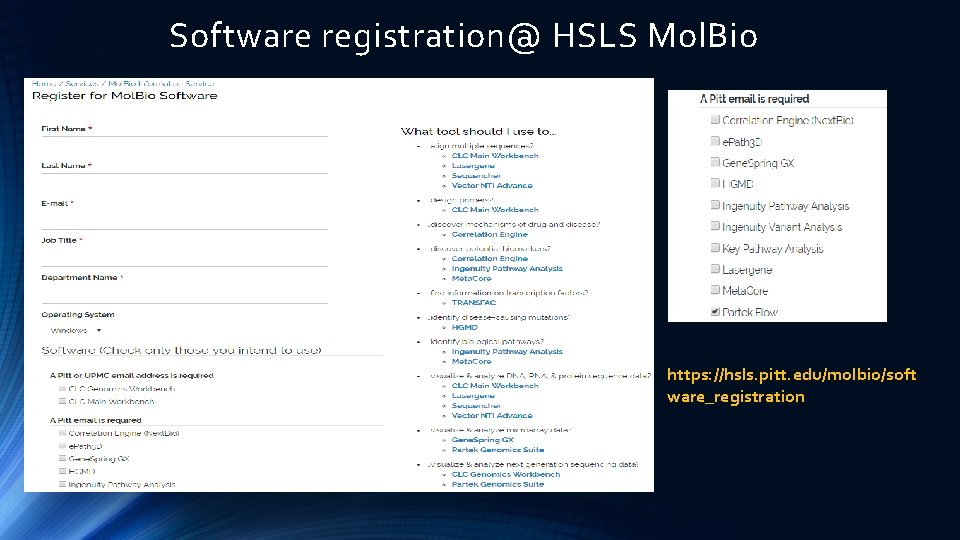
Software registration@ HSLS Mol. Bio https: //hsls. pitt. edu/molbio/soft ware_registration

Partek flow software access

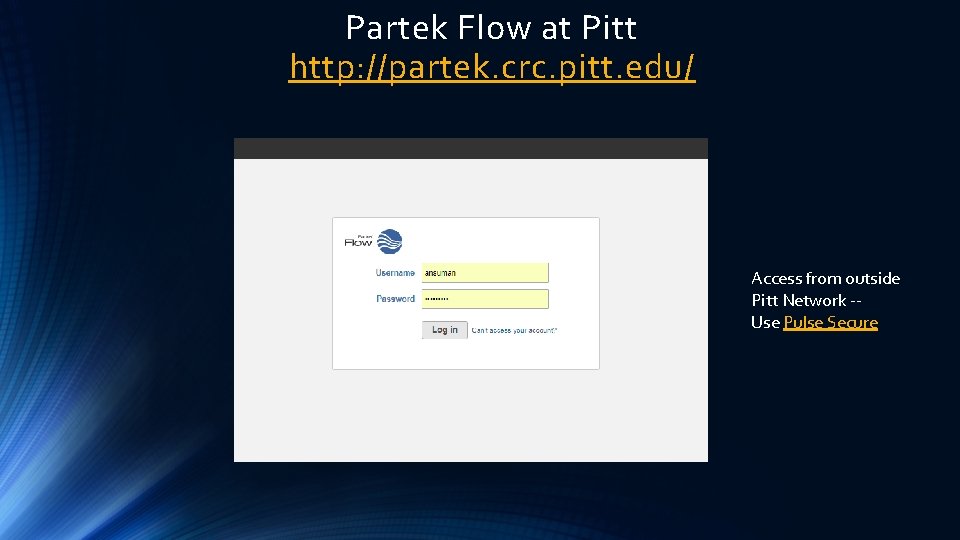
Partek Flow at Pitt http: //partek. crc. pitt. edu/ Access from outside Pitt Network -Use Pulse Secure

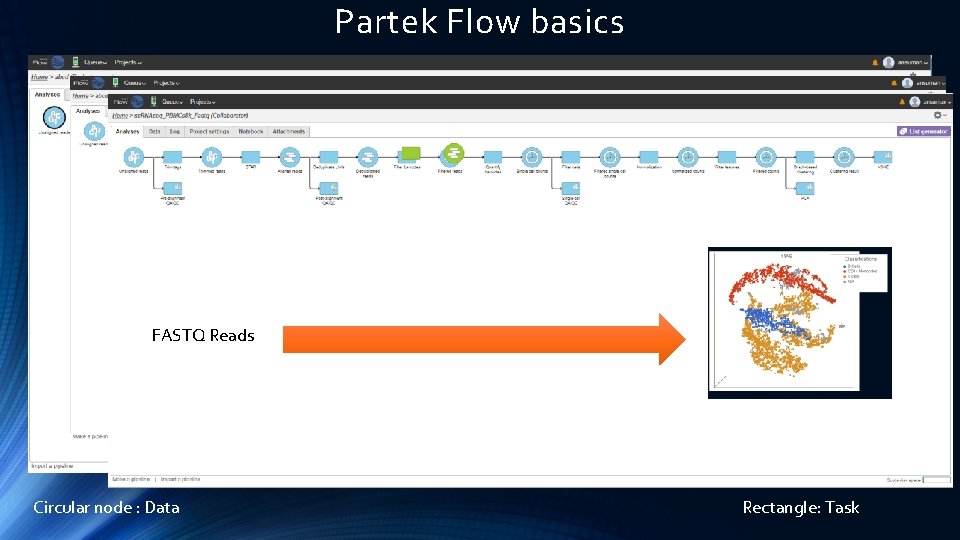
Partek Flow basics FASTQ Reads Circular node : Data Rectangle: Task

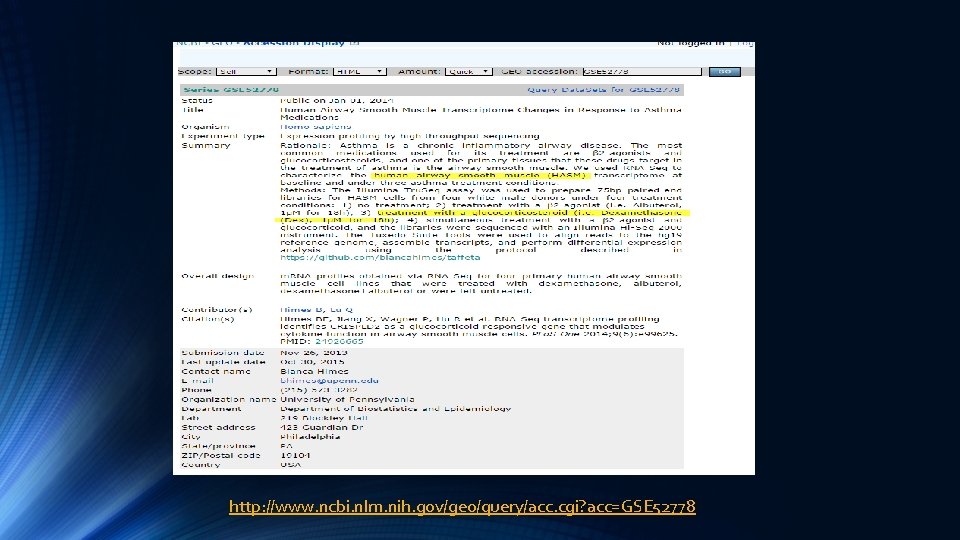
Bulk RNA-seq Study http: //journals. plos. org/plosone/article? id=10. 1371/journal. pone. 0099625

http: //www. ncbi. nlm. nih. gov/geo/query/acc. cgi? acc=GSE 52778

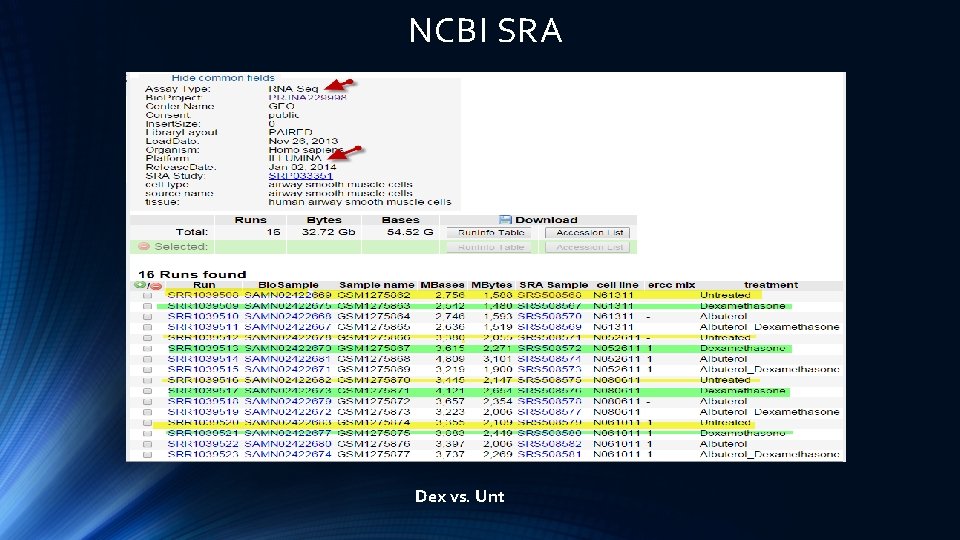
NCBI SRA Dex vs. Unt

Import CLCGx generated RNA-seq count matrix file into Partek. Flow

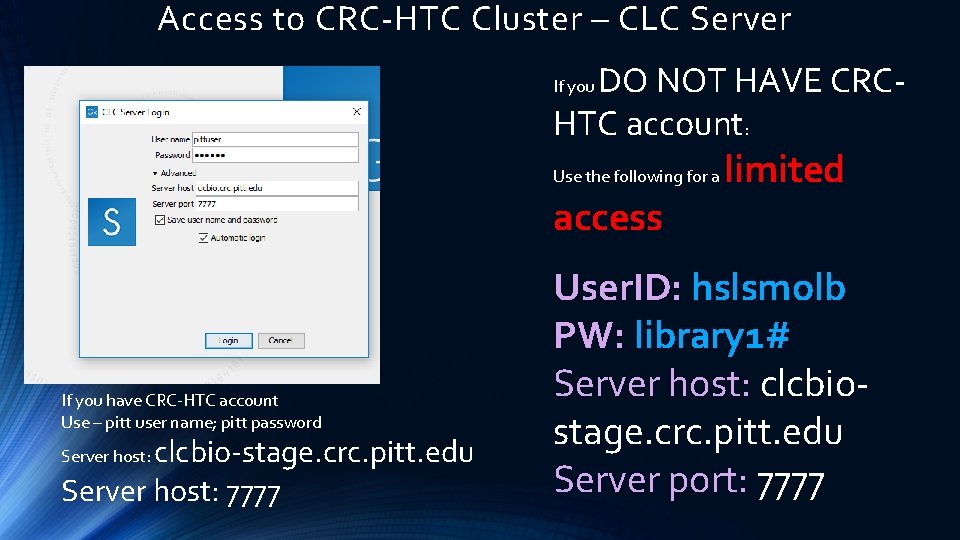
Access to CRC-HTC Cluster – CLC Server DO NOT HAVE CRCHTC account: If you Use the following for a access If you have CRC-HTC account Use – pitt user name; pitt password clcbio-stage. crc. pitt. edu Server host: 7777 Server host: limited User. ID: hslsmolb PW: library 1# Server host: clcbiostage. crc. pitt. edu Server port: 7777

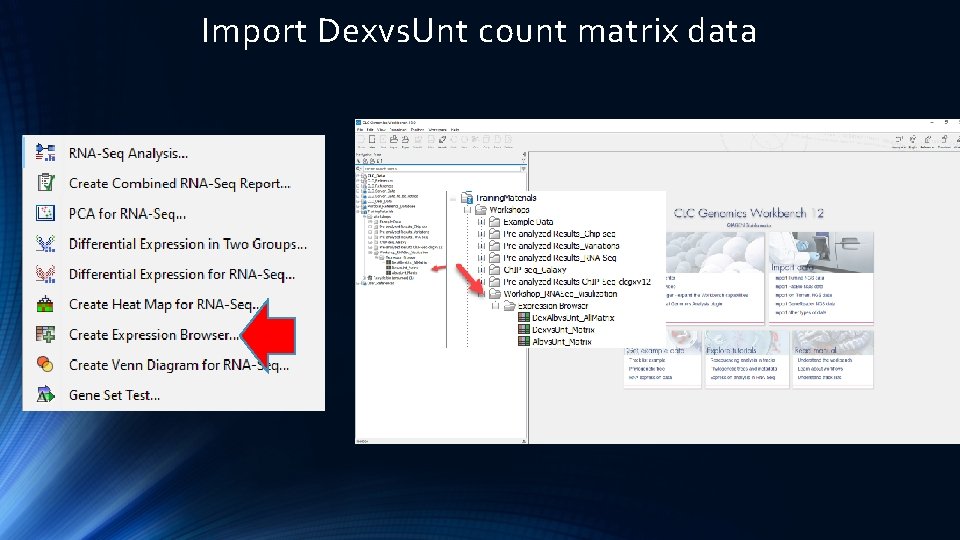
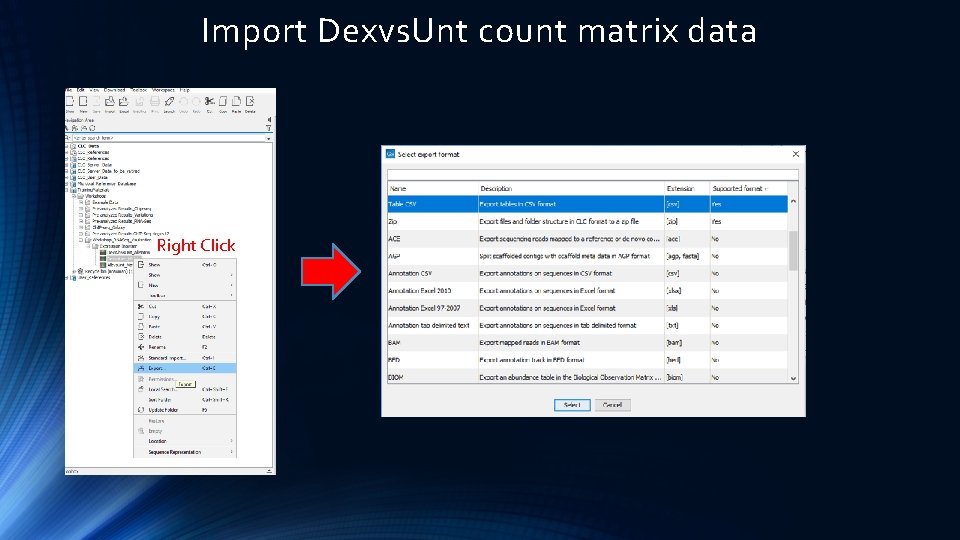
Import Dexvs. Unt count matrix data

Import Dexvs. Unt count matrix data Right Click

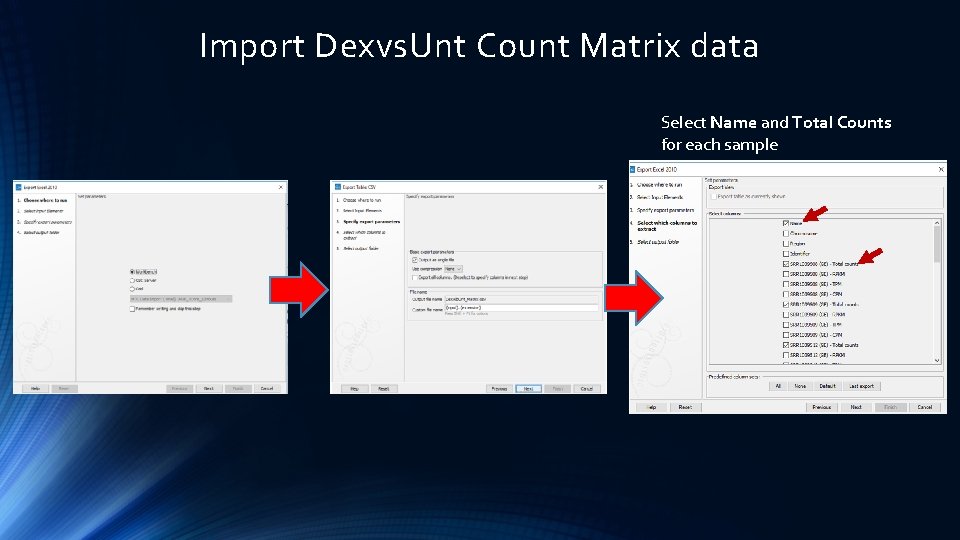
Import Dexvs. Unt Count Matrix data Select Name and Total Counts for each sample

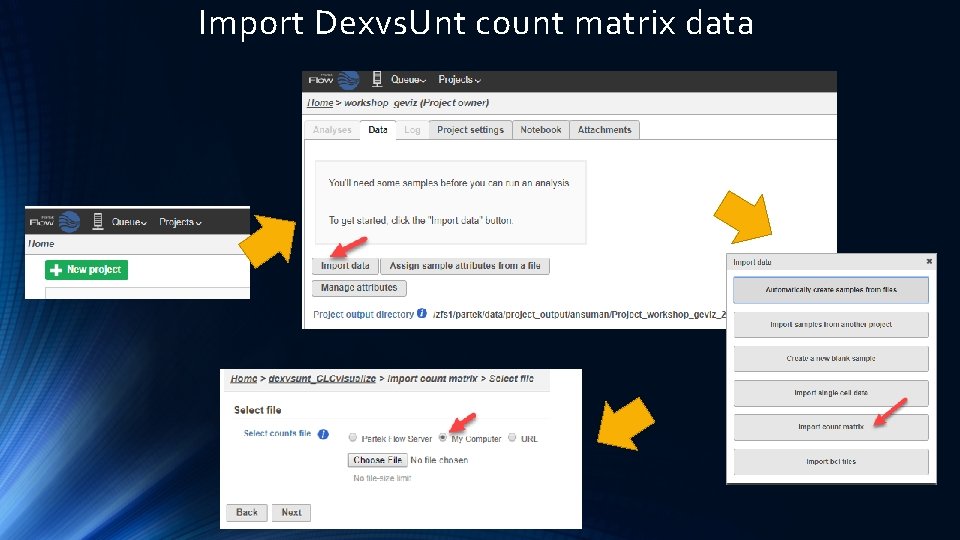
Import Dexvs. Unt count matrix data

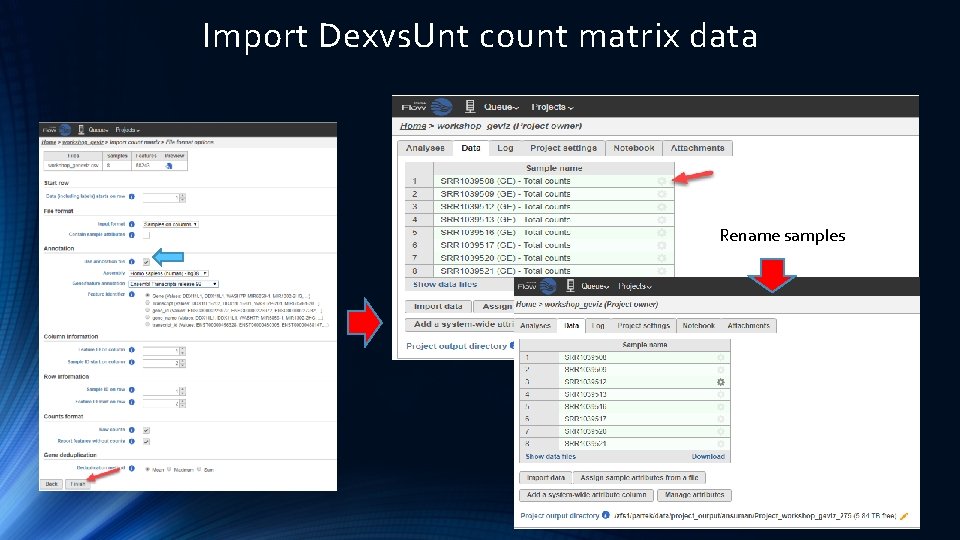
Import Dexvs. Unt count matrix data Rename samples

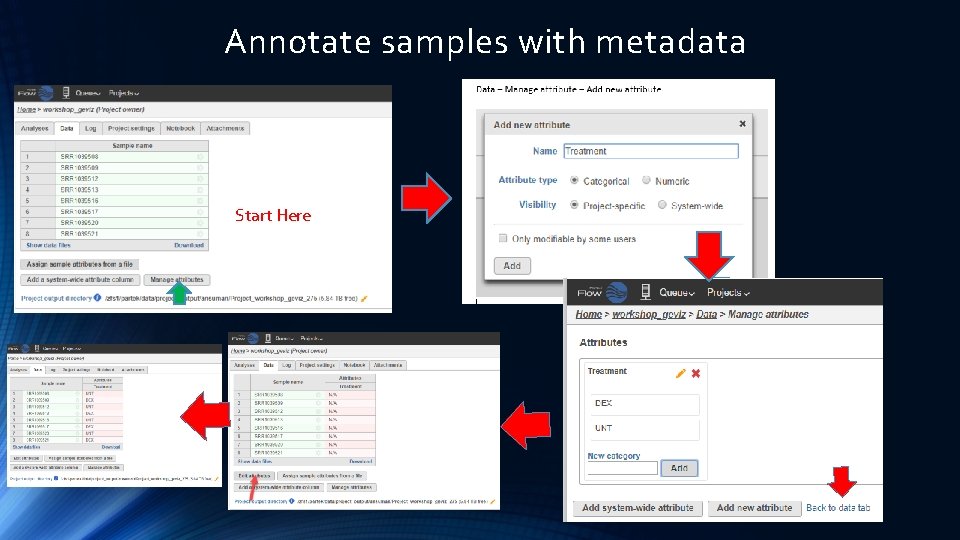
Annotate samples with metadata Start Here

Start analyzing the data and create visualization plots in Partek Flow

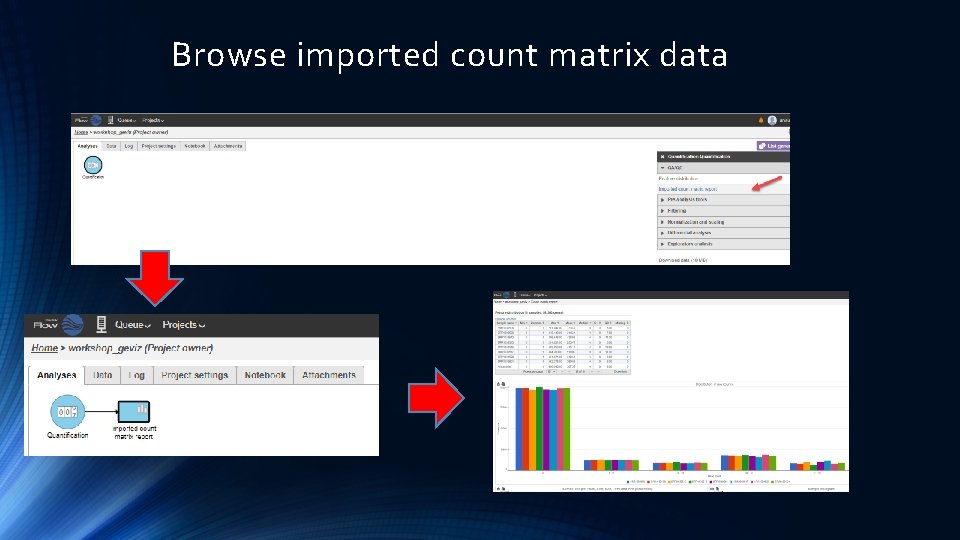
Browse imported count matrix data

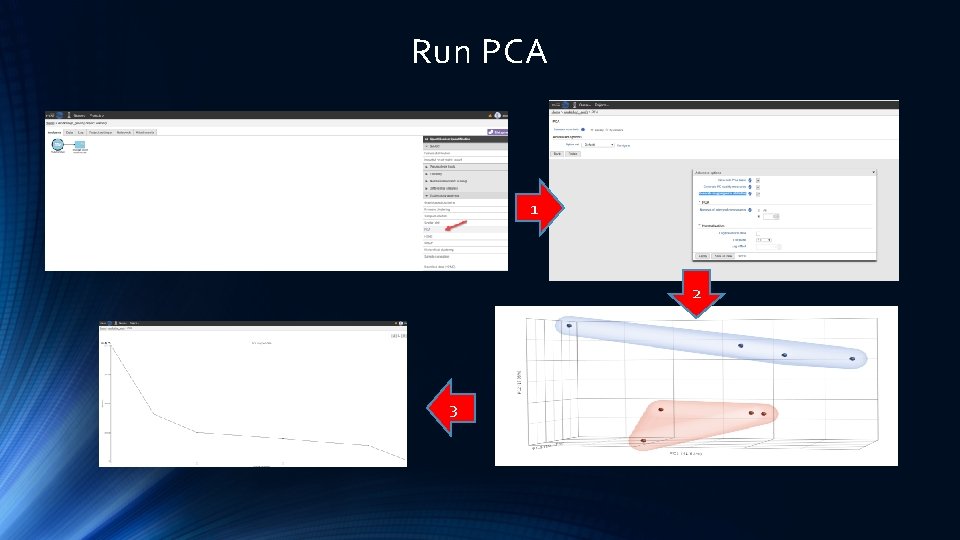
Run PCA 1 2 3

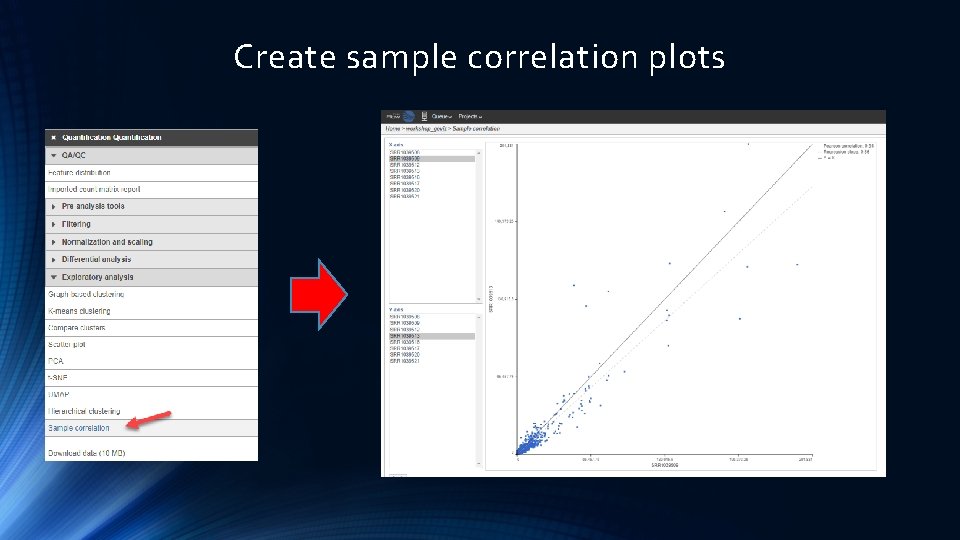
Create sample correlation plots

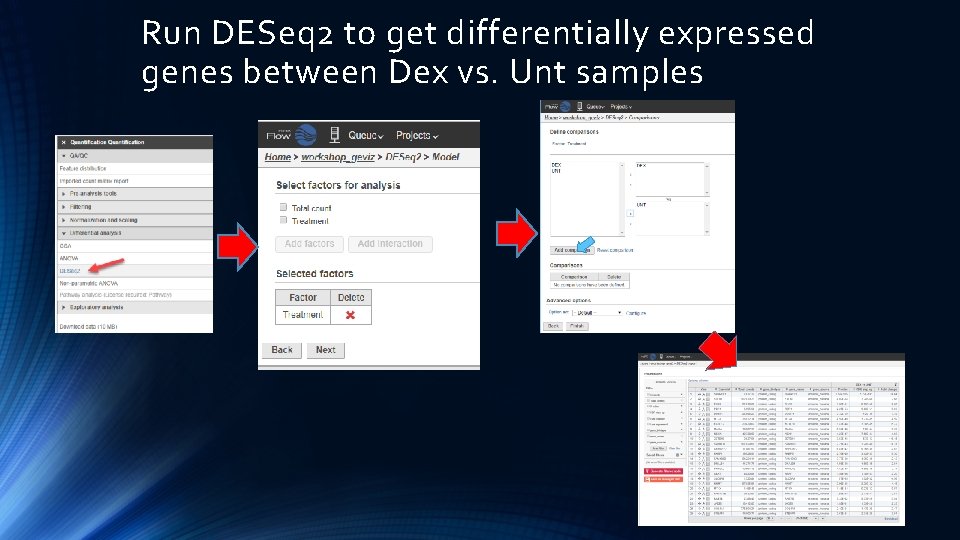
Run DESeq 2 to get differentially expressed genes between Dex vs. Unt samples

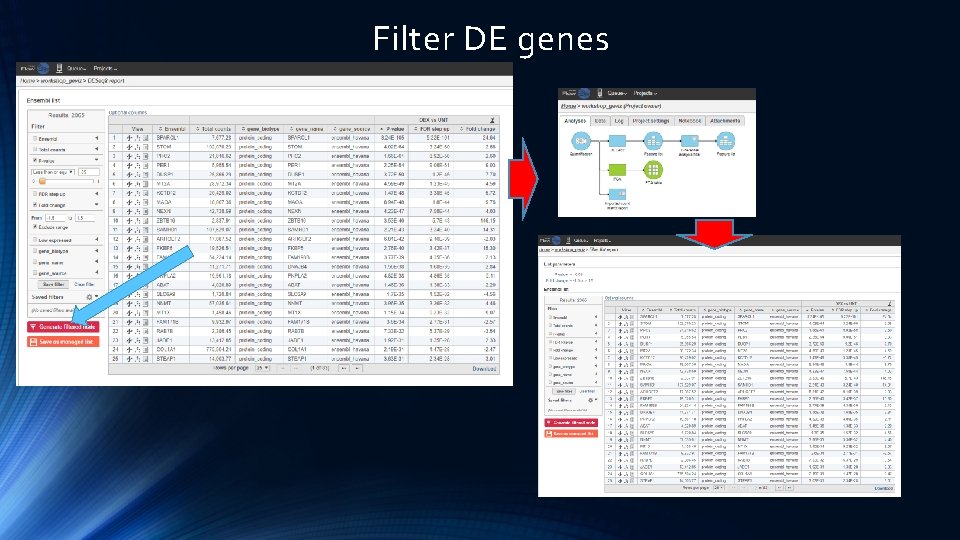
Filter DE genes

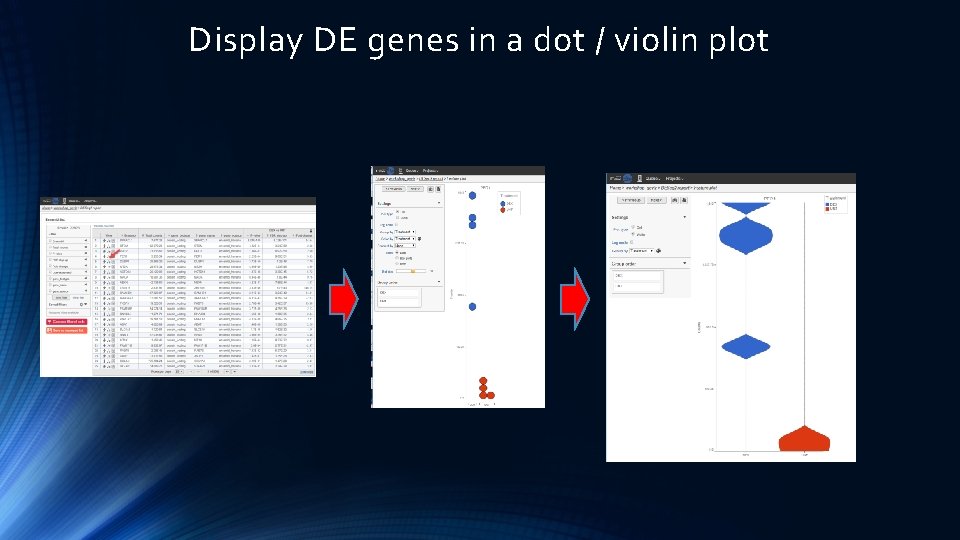
Display DE genes in a dot / violin plot

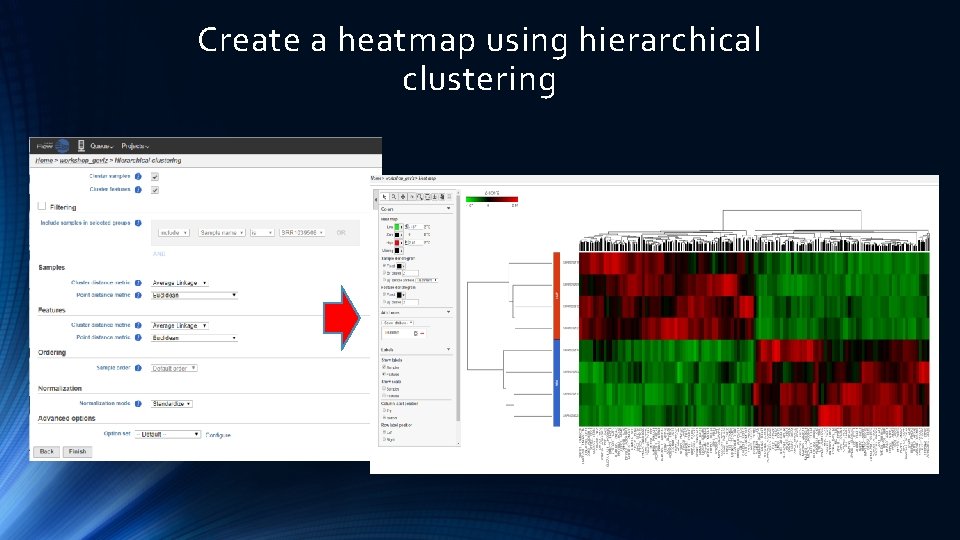
Create a heatmap using hierarchical clustering

Hands on exercise • Import “Alb vs. Unt” CLCGx created expression browser dataset into Partek Flow • Create a PCA Plot • Run DESeq 2 software and generate a differentially expressed gene list - Alb vs. Unt • Display DE genes in dot or violin plots • Create a heatmap displaying clustered samples in rows and clustered genes in columns • Create a Venn diagram showing overlap between DE genes (p-value <=. 05, FC <= -1. 5 and >=1. 5) produced by “Dex vs. Unt” and “Alb vs. Unt” datasets