The Statistics of Local Pairwise Sequence Alignment Part



- Slides: 18

The Statistics of Local Pairwise Sequence Alignment, Part I Stephen Altschul National Center for Biotechnology Information National Library of Medicine National Institutes of Health

Central Issues in Biological Sequence Comparison Definitions: What is one trying to find or optimize? Algorithms: Can one find or optimize the proposed object in reasonable time? Statistics: Can one’s result be explained by chance? In general there is a tension between these questions. A simple definition may allow efficient algorithms, but may not yield results of biological interest. However, a definition that includes most of the relevant biology may entail intractable algorithms and statistics. The most successful approaches find a balance between these considerations.

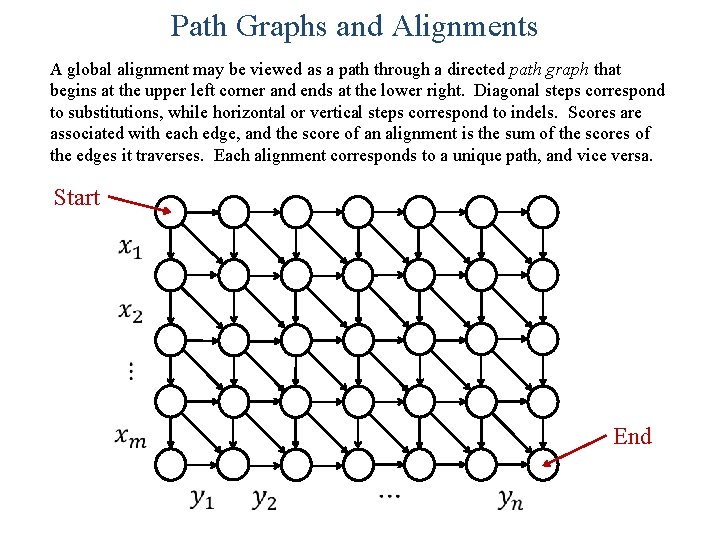
Path Graphs and Alignments A global alignment may be viewed as a path through a directed path graph that begins at the upper left corner and ends at the lower right. Diagonal steps correspond to substitutions, while horizontal or vertical steps correspond to indels. Scores are associated with each edge, and the score of an alignment is the sum of the scores of the edges it traverses. Each alignment corresponds to a unique path, and vice versa. Start End

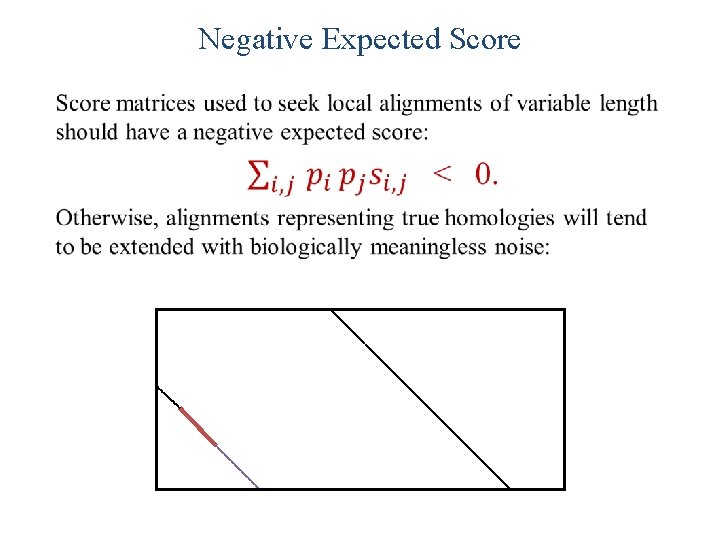
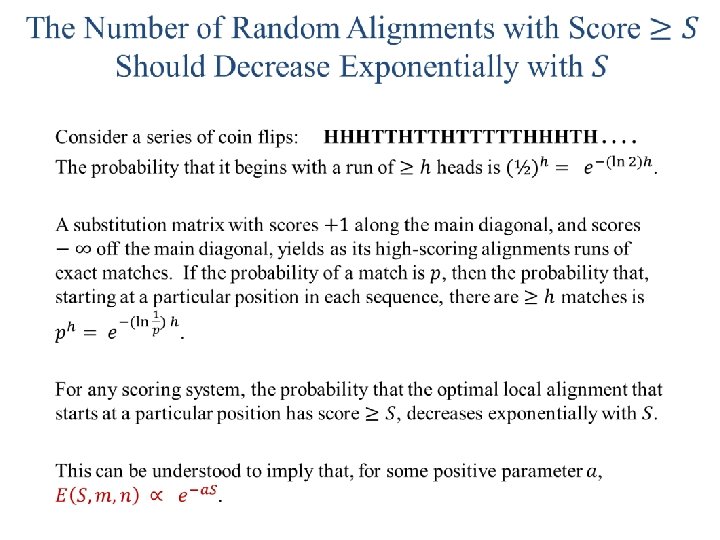
Ungapped Local Alignments When two sequences are compared, how great are the local alignment scores that can be expected to arise purely by chance? In other words, when can a local alignment be considered statistically significant? We will first develop the statistical theory for local alignments without gaps.

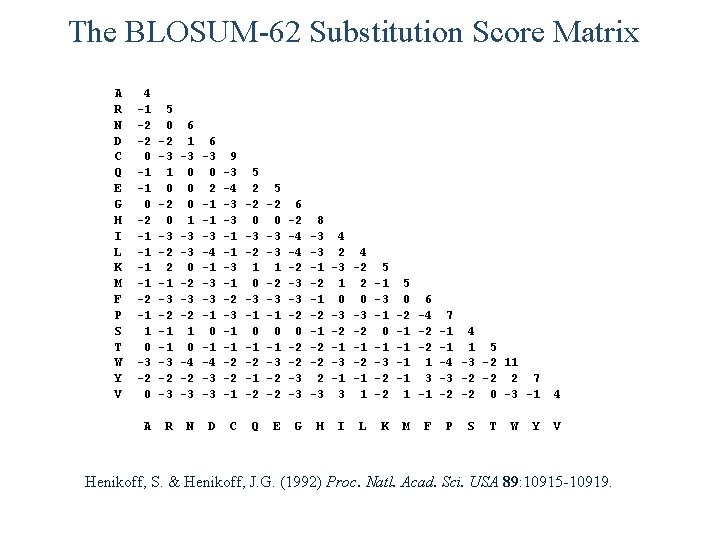
The BLOSUM-62 Substitution Score Matrix A R N D C Q E G H I L K M F P S T W Y V 4 -1 -2 -2 0 -1 -1 0 -2 -1 -1 -2 -1 1 0 -3 -2 0 5 0 -2 -3 1 0 -2 0 -3 -2 2 -1 -3 -2 -1 -1 -3 -2 -3 6 1 -3 0 0 0 1 -3 -3 0 -2 -3 -2 1 0 -4 -2 -3 6 -3 0 2 -1 -1 -3 -4 -1 -3 -3 -1 0 -1 -4 -3 -3 9 -3 -4 -3 -3 -1 -1 -3 -1 -2 -3 -1 -1 -2 -2 -1 5 2 -2 0 -3 -2 1 0 -3 -1 0 -1 -2 5 -2 0 -3 -3 1 -2 -3 -1 0 -1 -3 -2 -2 6 -2 -4 -4 -2 -3 -3 -2 0 -2 -2 -3 -3 8 -3 -3 -1 -2 -2 2 -3 4 2 -3 1 0 -3 -2 -1 -3 -1 3 4 -2 2 0 -3 -2 -1 1 5 -1 -3 -1 0 -1 -3 -2 -2 5 0 -2 -1 -1 1 6 -4 -2 -2 1 3 -1 A R N D C Q E G H I L K M F 7 -1 4 -1 1 5 -4 -3 -2 11 -3 -2 -2 2 7 -2 -2 0 -3 -1 P S T W Y 4 V Henikoff, S. & Henikoff, J. G. (1992) Proc. Natl. Acad. Sci. USA 89: 10915 -10919.

Negative Expected Score •

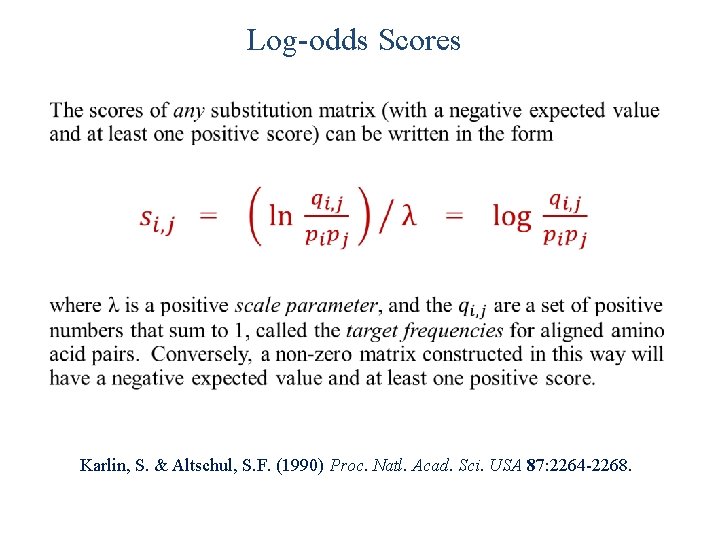
Log-odds Scores • Karlin, S. & Altschul, S. F. (1990) Proc. Natl. Acad. Sci. USA 87: 2264 -2268.

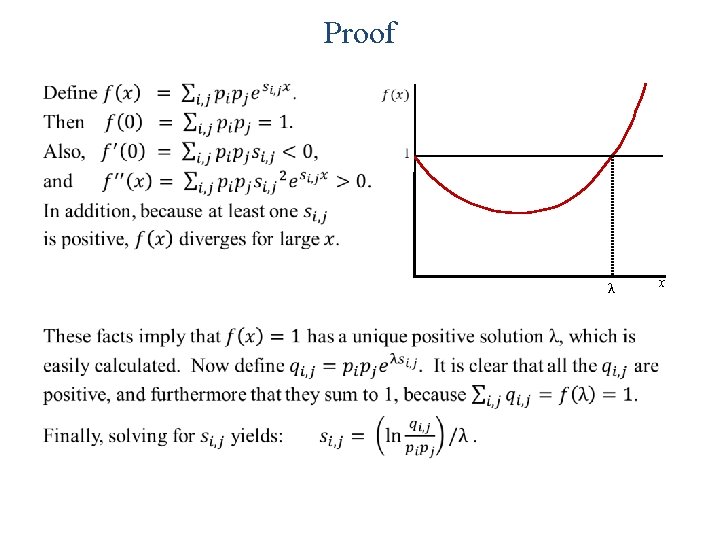
Proof • λ x

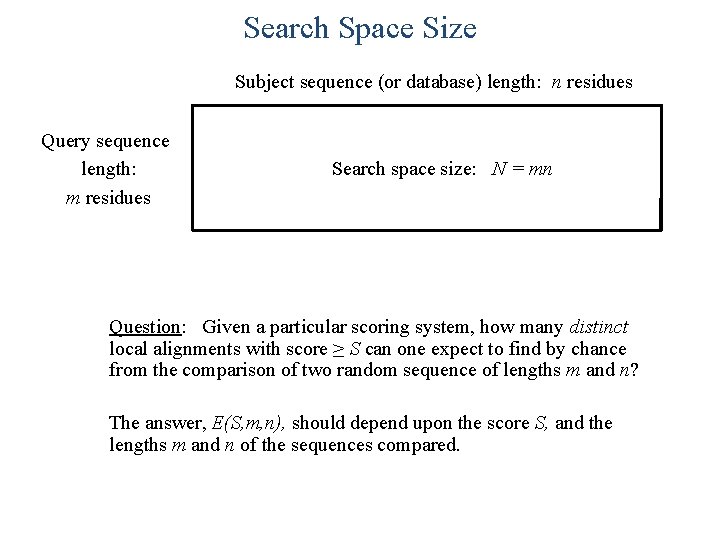
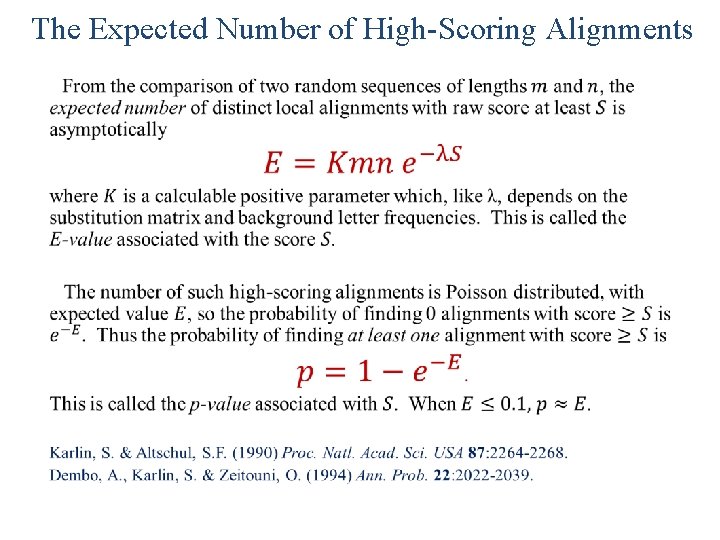
Search Space Size Subject sequence (or database) length: n residues Query sequence length: m residues Search space size: N = mn Question: Given a particular scoring system, how many distinct local alignments with score ≥ S can one expect to find by chance from the comparison of two random sequence of lengths m and n? The answer, E(S, m, n), should depend upon the score S, and the lengths m and n of the sequences compared.

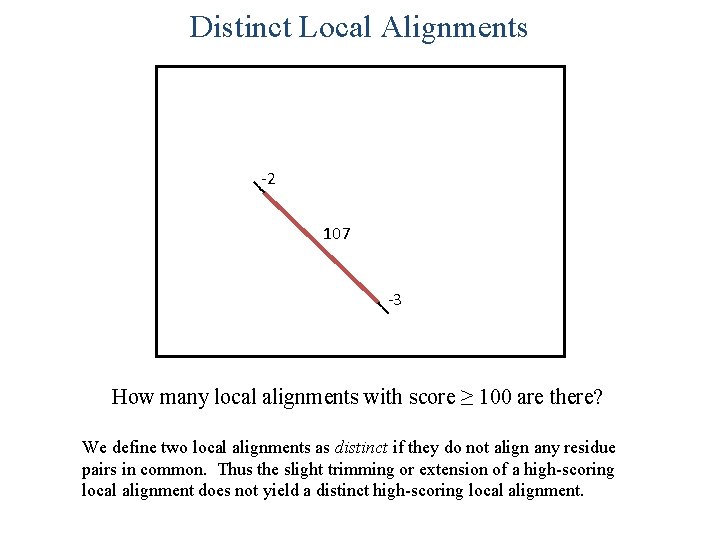
Distinct Local Alignments -2 107 -3 How many local alignments with score ≥ 100 are there? We define two local alignments as distinct if they do not align any residue pairs in common. Thus the slight trimming or extension of a high-scoring local alignment does not yield a distinct high-scoring local alignment.

The Number of Random High-scoring Alignments Should be Proportional to the Search Space Size •


The Expected Number of High-Scoring Alignments •

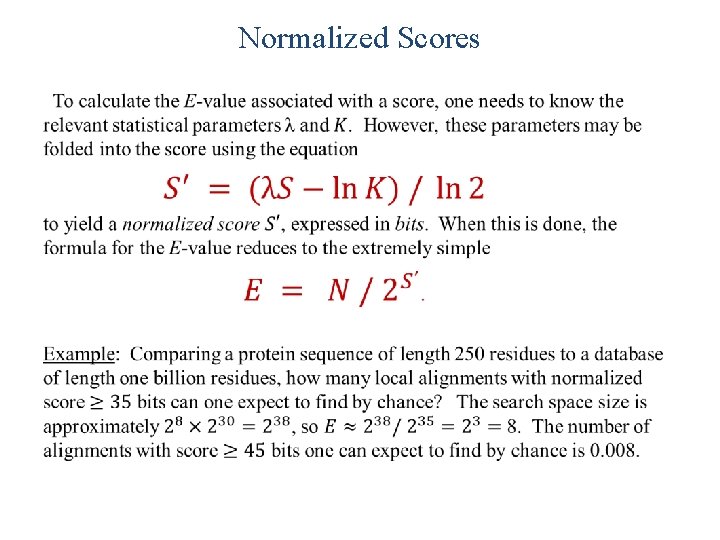
Normalized Scores •

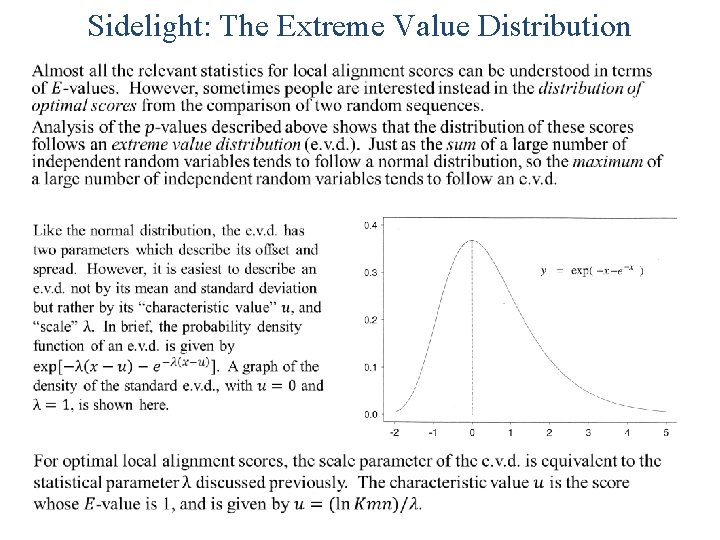
Sidelight: The Extreme Value Distribution •

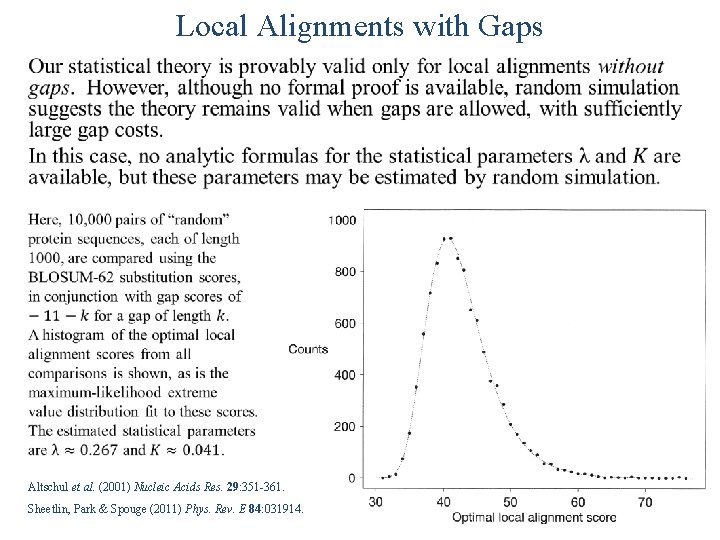
Local Alignments with Gaps Altschul et al. (2001) Nucleic Acids Res. 29: 351 -361. Sheetlin, Park & Spouge (2011) Phys. Rev. E 84: 031914.

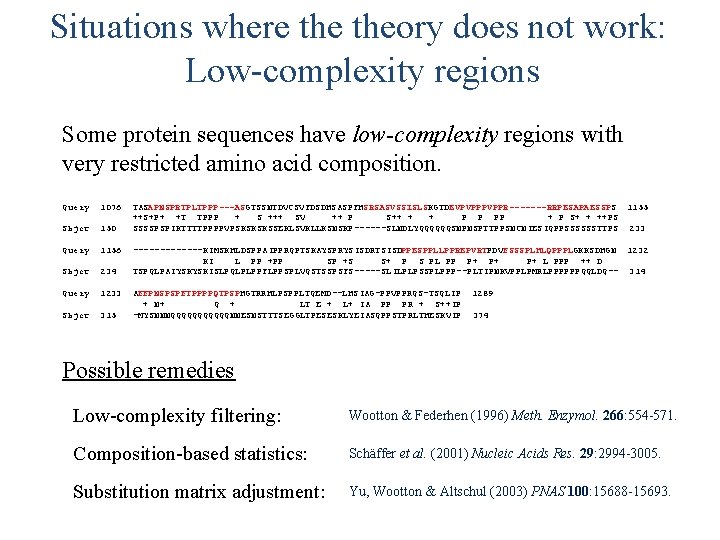
Situations where theory does not work: Low-complexity regions Some protein sequences have low-complexity regions with very restricted amino acid composition. Query 1076 Sbjct 150 Query 1156 Sbjct 234 Query 1233 Sbjct 315 TASAPNSPRTPLTPPP---ASGTSSNTDVCSVFDSDHSASPFHSRSASVSSISLSKGTDEVPVPPR-------RRPESAPAESSPS 1155 ++S+P+ +T TPPP + S +++ SV ++ P S++ + + P P PP + P S+ + ++PS SSSSPSPIKTTTTPPPPVPSKSKSKSSEKLSVKLLKSNSKP------SLNDLYQQQQQQSNPNSPTTPPSNCNIESIQPPSSSSSSTTPS 233 -------KIMSKHLDSPPAIPPRQPTSKAYSPRYSISDRTSISDPPESPPLLPPREPVRTPDVFSSSPLHLQPPPLGKKSDHGN 1232 KI L PP +PP SP +S S+ P S PL PP P+ P+ P+ L PPP ++ D TSPQLPAIYSKYSKISLPQLPLPPFLPPSPLVQSTSSPSFS-----SLILPLPSSPLPPP--PLTIPNKVPPLPMRLPPPPPPQQLDQ-- 314 AFFPNSPSPFTPPPPQTPSPHGTRRHLPSPPLTQEMD--LHSIAG-PPVPPRQS-TSQLIP + N+ Q + LT E + L+ IA PP PR + S++IP -MYSNNNQQQQQQNNESNSTTTSEGGLTPESESKLYEIASQPPSTPRLTHESKVIP 1289 374 Possible remedies Low-complexity filtering: Wootton & Federhen (1996) Meth. Enzymol. 266: 554 -571. Composition-based statistics: Schäffer et al. (2001) Nucleic Acids Res. 29: 2994 -3005. Substitution matrix adjustment: Yu, Wootton & Altschul (2003) PNAS 100: 15688 -15693.

Situations where theory does not work: Violated asymptotics The expected length of alignments required to achieve a large score S may approach or exceed the lengths of one or both of the sequences being compared. When this is the case, the asymptotic theory described above breaks down. However, “edge effect” corrections to theory are available: Altschul, S. F. , et al. (2001) Nucleic Acids Res. 29: 351 -361. Park, Y. , et al. (2012) BMC Research Notes 5: 286.