software of life Genomes to function Lessons from

























- Slides: 55

“software” of life

Genomes to function

Lessons from genome projects • Most genes have no known function • Most genes w/ known function assigned from sequence-similarity matches to other organisms • Need methods to experimentally assay gene activity on a genome-wide scale

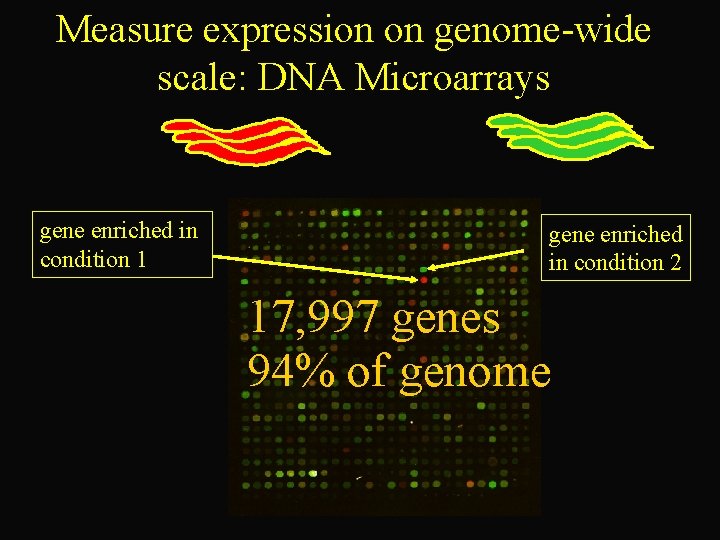
Measure expression on genome-wide scale: DNA Microarrays Condition 1 RNA gene enriched in condition 1 Condition 2 RNA gene enriched in condition 2 17, 997 genes 94% of genome

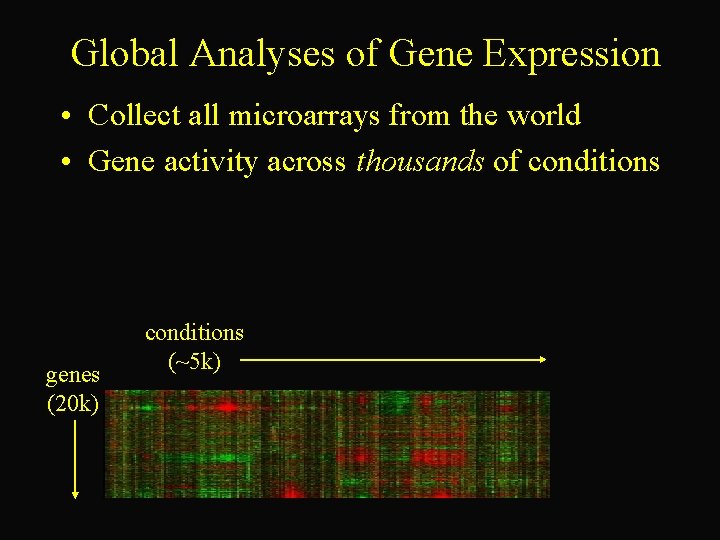
Global Analyses of Gene Expression • Collect all microarrays from the world • Gene activity across thousands of conditions genes (20 k) conditions (~5 k)

Digital Age of Biology • Biologists drowning in data • Bottleneck now is developing computational resources for discovery • Think Genbank before BLAST. . .

Discovering Gene Function on a Global Scale • Gene Networks • Search Engines

Gene Networks Matt Weirauch. Martina Koeva Corey Powell Charlie Vaske Alex Chad Williams Chen

Gene Networks • link 2 genes together if they are co-activated in multiple organisms • build networks from all the links • discover function from a gene’s links • understand bigger picture of gene regulation

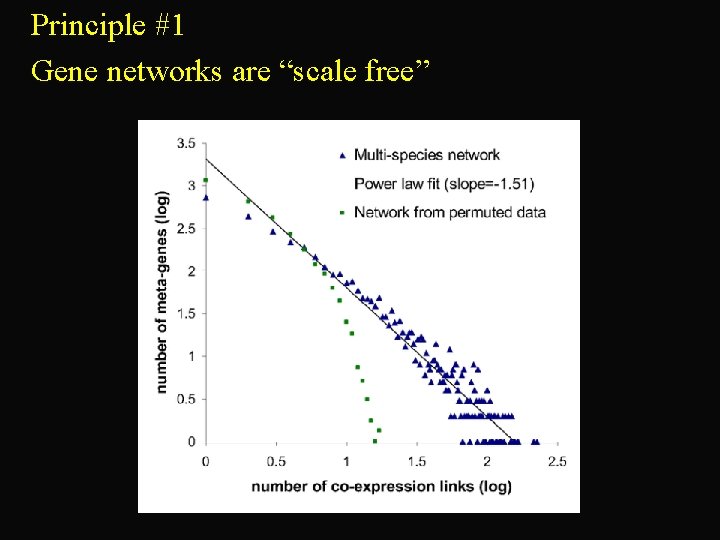
Principle #1 Gene networks are “scale free”

• Scale free – gene networks may arise from processes like expansion of WWW some links on the WWW

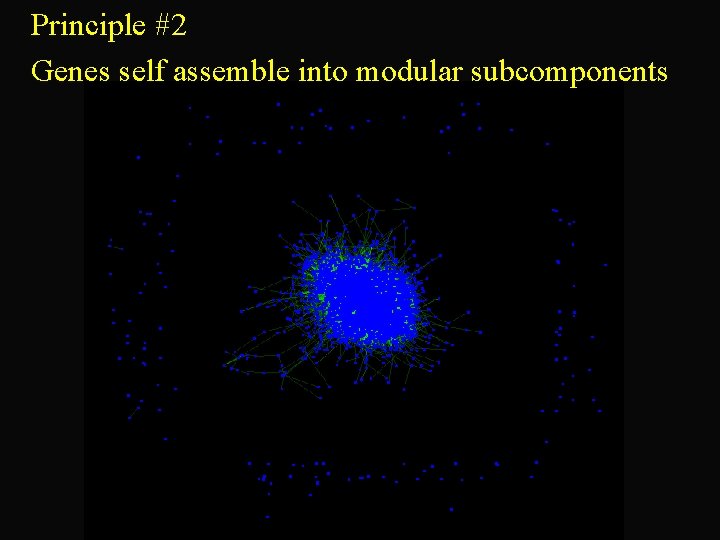
Principle #2 Genes self assemble into modular subcomponents

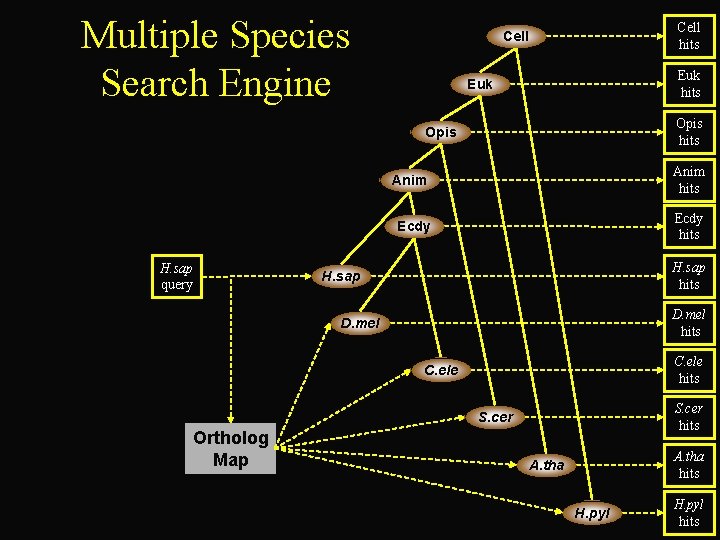
http: //www. cse. ucsc. edu/~jstuart/multispecies

Principle #2 Genes self assemble into modular subcomponents

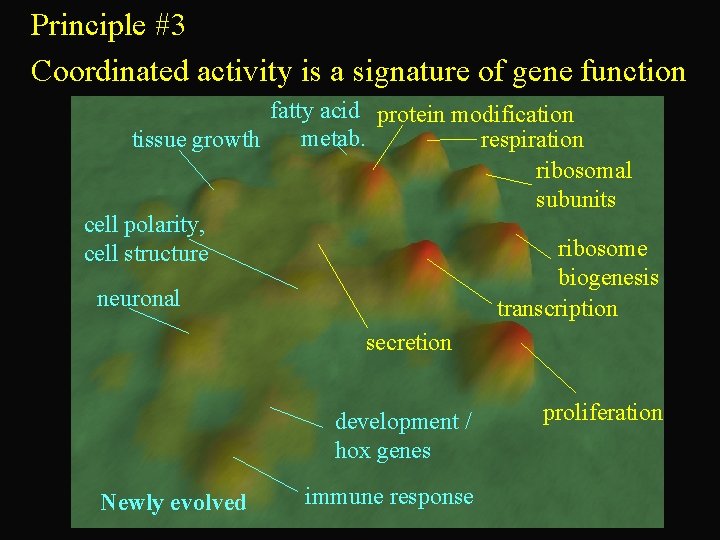
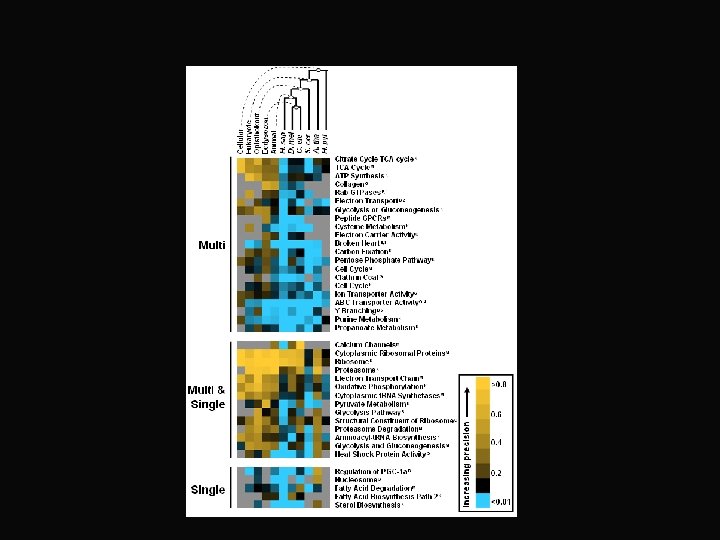
Principle #3 Coordinated activity is a signature of gene function fatty acid protein modification metab. respiration tissue growth ribosomal subunits cell polarity, ribosome cell structure biogenesis neuronal transcription secretion development / hox genes Newly evolved immune response proliferation

Proteasome “module” http: //www. cse. ucsc. edu/~jstuart/multispecies

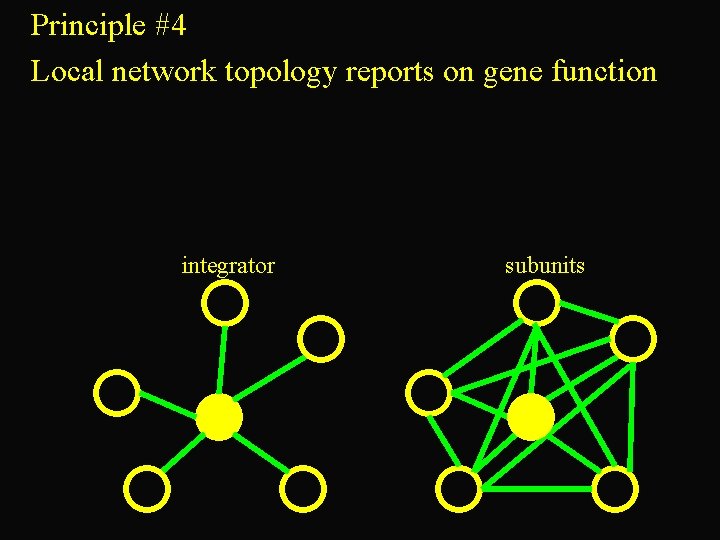
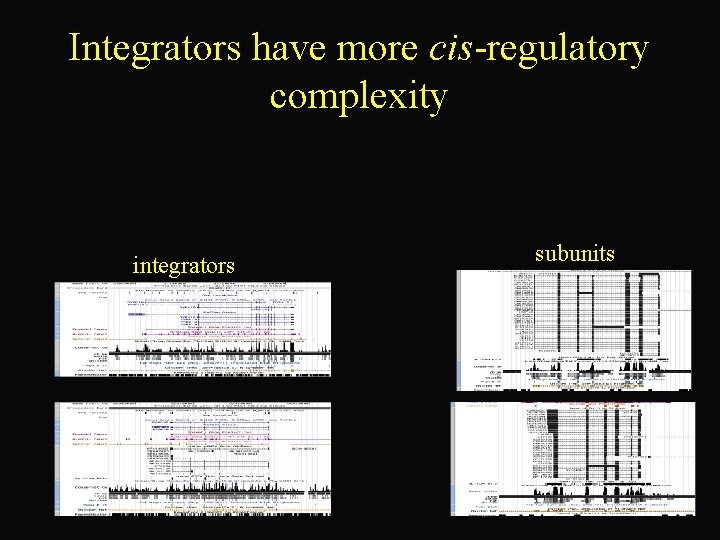
Principle #4 Local network topology reports on gene function integrator subunits

top 3 integrators:

Integrators have more cis-regulatory complexity integrators subunits

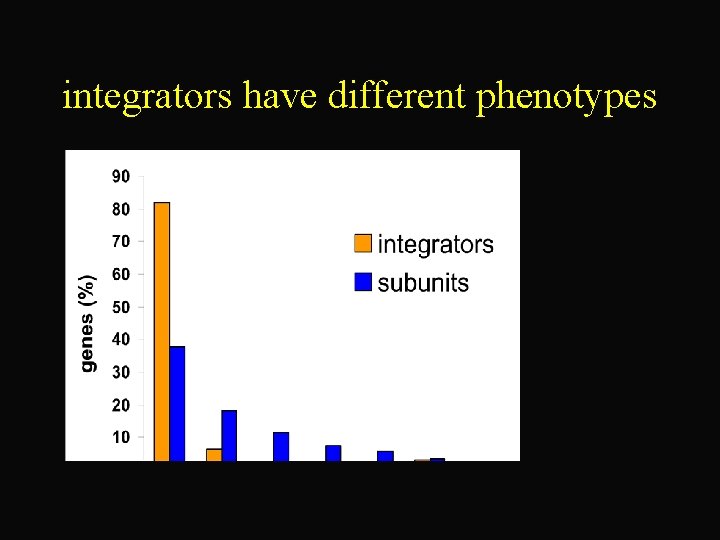
integrators have different phenotypes

Current Directions for Gene Networks • Gene isoform networks to capture alternative splicing • Predict drug targets from synthetic lethal nets

Current Directions for Gene Networks • Gene isoform networks to capture alternative splicing • Predict drug targets from synthetic lethal nets (w/ Lokey Lab)

Gene Isoform Networks Matt Weirauch. Martina Koeva Corey Powell Charlie Vaske Alex Chad Williams Chen

Gene Isoform Networks • Most human genes (>60%) are alternatively spliced. • Alternative splicing gives rise to different proteins from the same gene • The particular variant expressed can be very important (e. g. sex determination in flies) • The functional implications of alt. splicing in humans is still largely unexplored. • Provides a higher resolution understanding of gene expression and its relationship to health & disease

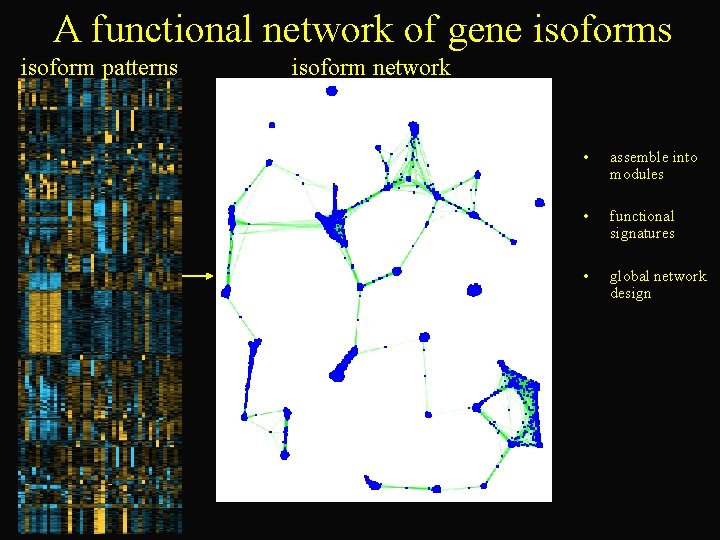
Splicing Microarrays • Measure particular subparts of the gene structure (e. g. exon-exon junctions) • Data now available for human and mouse tissue compendiums • Infer isoforms from expression of subparts across the tissues • Identify isoform modules

A functional network of gene isoforms isoform patterns isoform network • assemble into modules • functional signatures • global network design

Search Engines Matt Weirauch. Martina Koeva Corey Powell Charlie Vaske Alex Chad Williams Chen

Search engines to discover gene function

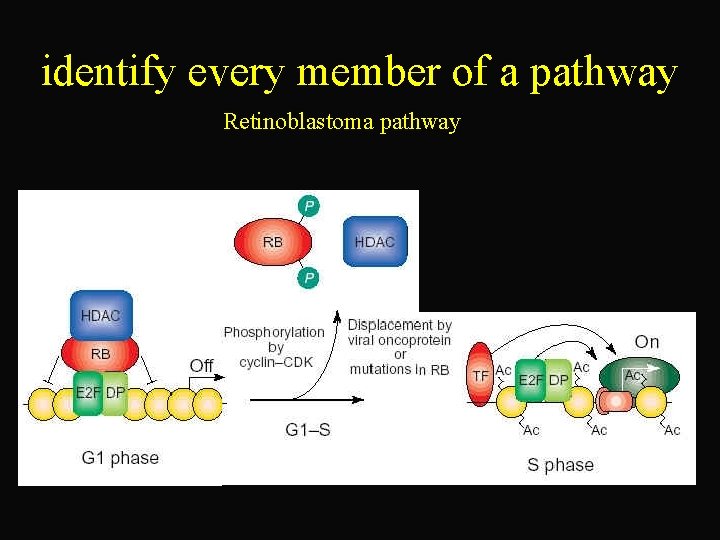
identify every member of a pathway Retinoblastoma pathway

(slide from Art Owen)

gene recommender search for regulating conditions query

gene recommender search for regulating conditions query

gene recommender regulating conditions query search for new candidates

gene recommender regulating conditions query + “hits”

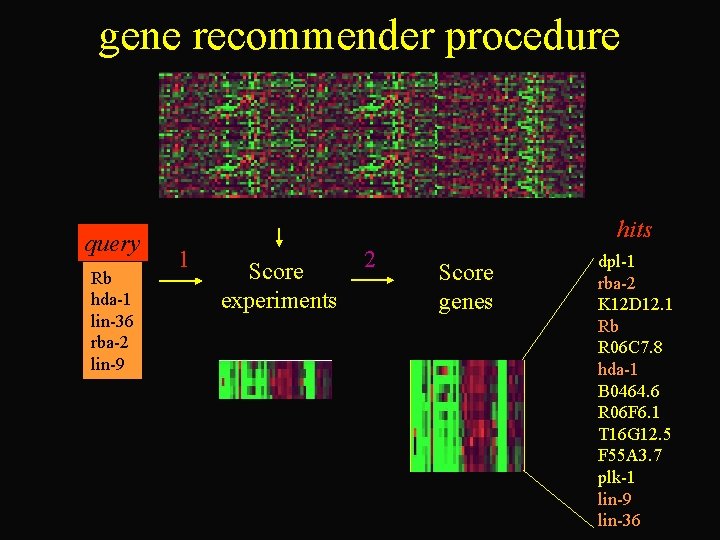
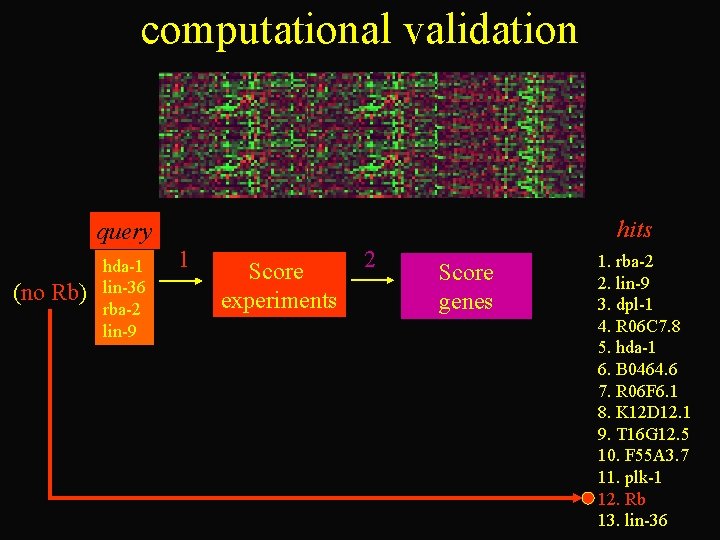
gene recommender procedure query Rb hda-1 lin-36 rba-2 lin-9 hits 1 Score experiments 2 Score genes dpl-1 rba-2 K 12 D 12. 1 Rb R 06 C 7. 8 hda-1 B 0464. 6 R 06 F 6. 1 T 16 G 12. 5 F 55 A 3. 7 plk-1 lin-9 lin-36

computational validation hits query (no Rb) hda-1 lin-36 rba-2 lin-9 1 Score experiments 2 Score genes 1. rba-2 2. lin-9 3. dpl-1 4. R 06 C 7. 8 5. hda-1 6. B 0464. 6 7. R 06 F 6. 1 8. K 12 D 12. 1 9. T 16 G 12. 5 10. F 55 A 3. 7 11. plk-1 12. Rb 13. lin-36

Searching 1 organism

Multiple Species Search Engine Cell hits Cell Euk hits Euk Opis hits Opis H. sap query Anim hits Ecdy hits H. sap D. mel hits D. mel C. ele hits C. ele S. cer hits S. cer Ortholog Map A. tha hits A. tha H. pyl hits

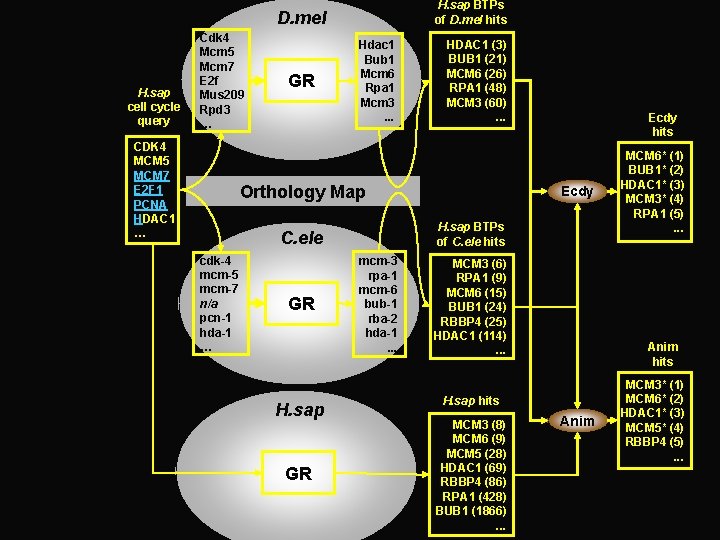
H. sap BTPs of D. mel hits D. mel H. sap cell cycle query Cdk 4 Mcm 5 Mcm 7 E 2 f Mus 209 Rpd 3 … CDK 4 MCM 5 MCM 7 E 2 F 1 PCNA HDAC 1 … GR Hdac 1 Bub 1 Mcm 6 Rpa 1 Mcm 3. . . HDAC 1 (3) BUB 1 (21) MCM 6 (26) RPA 1 (48) MCM 3 (60). . . Orthology Map GR H. sap GR Ecdy H. sap BTPs of C. ele hits C. ele cdk-4 mcm-5 mcm-7 n/a pcn-1 hda-1 … Ecdy hits mcm-3 rpa-1 mcm-6 bub-1 rba-2 hda-1. . . MCM 3 (6) RPA 1 (9) MCM 6 (15) BUB 1 (24) RBBP 4 (25) HDAC 1 (114). . . Anim hits H. sap hits MCM 3 (8) MCM 6 (9) MCM 5 (28) HDAC 1 (69) RBBP 4 (86) RPA 1 (428) BUB 1 (1866). . . MCM 6* (1) BUB 1* (2) HDAC 1* (3) MCM 3* (4) RPA 1 (5). . . Anim MCM 3* (1) MCM 6* (2) HDAC 1* (3) MCM 5* (4) RBBP 4 (5). . .

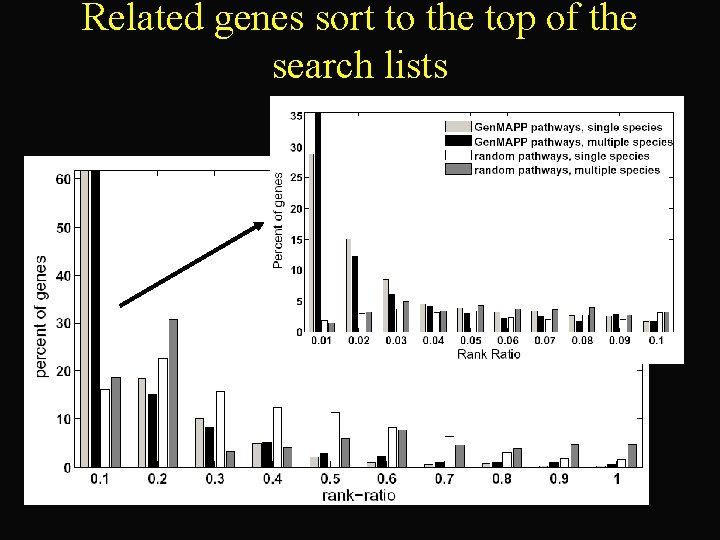
Related genes sort to the top of the search lists

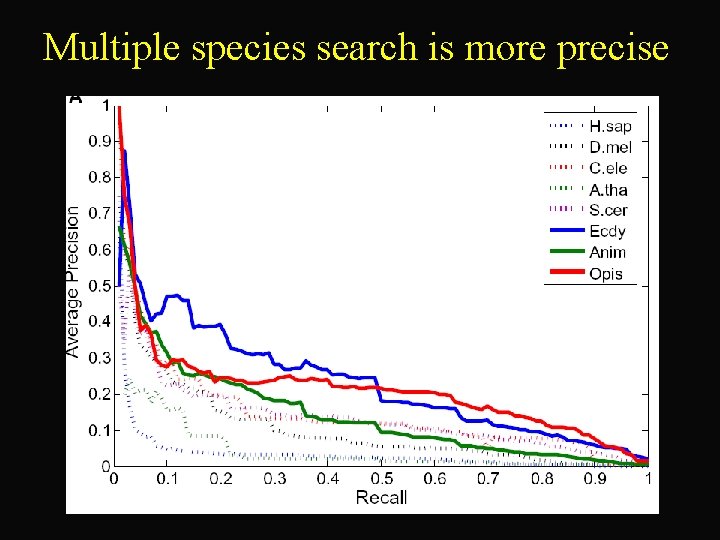
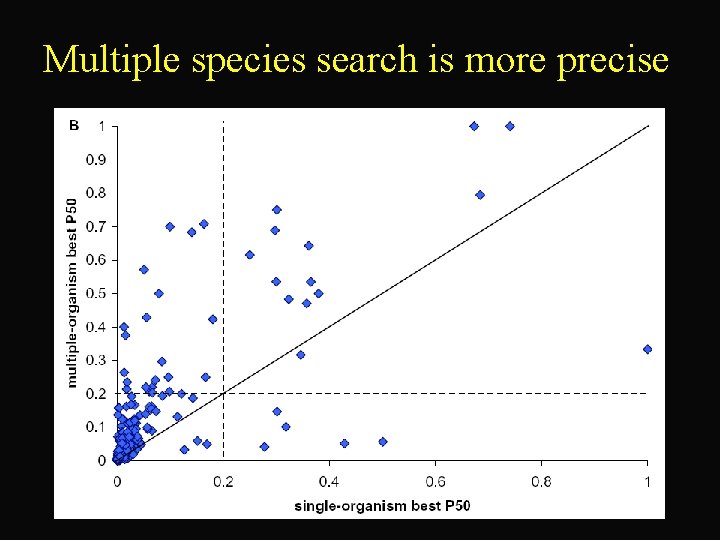
Multiple species search is more precise

Multiple species search is more precise


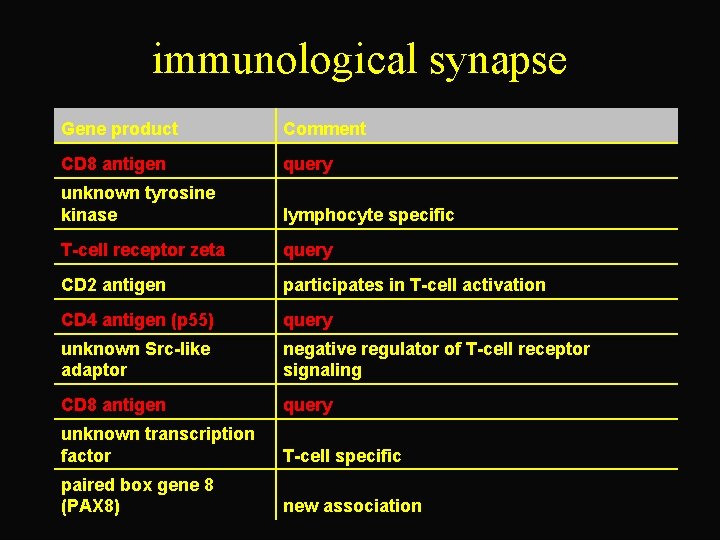
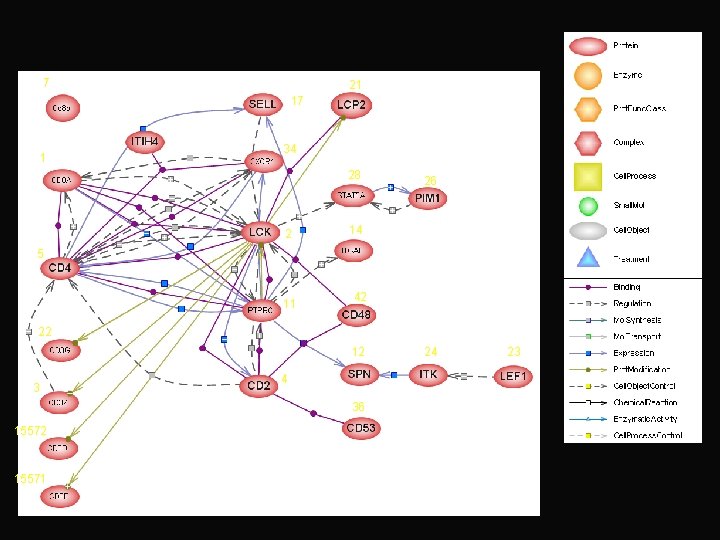
immunological synapse Gene product Comment CD 8 antigen query unknown tyrosine kinase lymphocyte specific T-cell receptor zeta query CD 2 antigen participates in T-cell activation CD 4 antigen (p 55) query unknown Src-like adaptor negative regulator of T-cell receptor signaling CD 8 antigen query unknown transcription factor T-cell specific paired box gene 8 (PAX 8) new association

7 21 17 1 34 28 2 26 14 5 11 42 22 12 3 4 36 15572 15571 24 23

Search Engine Directions • Search gene networks for pathway members – Incorporate multiple data sources in search – Faster than scanning raw data • Discriminative search engines – E. g. identify genes coregulated with DNA damage genes more so than S-phase genes

Search Engine Directions • Network Recommender – Search gene networks for pathway members – Incorporate multiple data sources in search – Faster than scanning raw data • Discriminative search engines – E. g. identify genes coregulated with DNA damage genes more so than S-phase genes

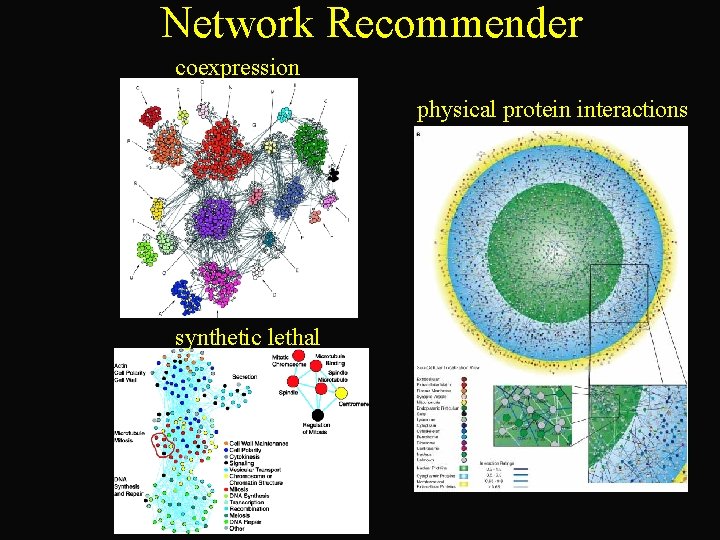
Network Recommender Matt Weirauch. Martina Koeva Corey Powell Charlie Vaske Alex Chad Williams Chen

Network Recommender coexpression physical protein interactions synthetic lethal

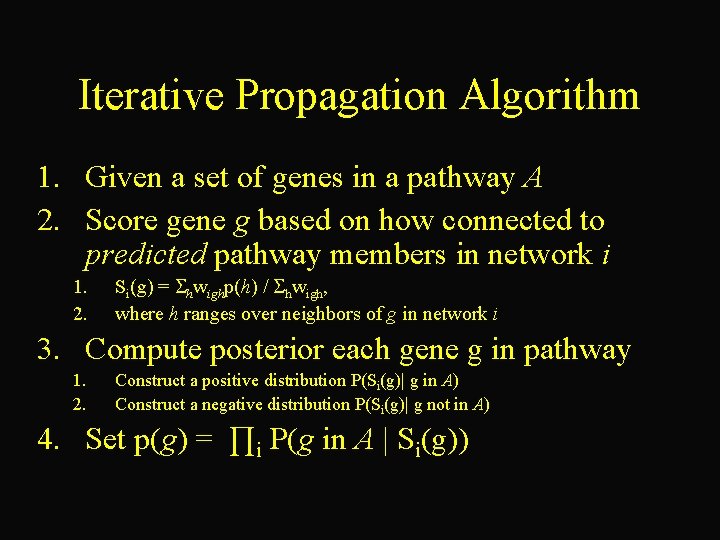
Iterative Propagation Algorithm 1. Given a set of genes in a pathway A 2. Score gene g based on how connected to predicted pathway members in network i 1. 2. Si(g) = hwighp(h) / hwigh, where h ranges over neighbors of g in network i 3. Compute posterior each gene g in pathway 1. 2. Construct a positive distribution P(Si(g)| g in A) Construct a negative distribution P(Si(g)| g not in A) 4. Set p(g) = ∏i P(g in A | Si(g))

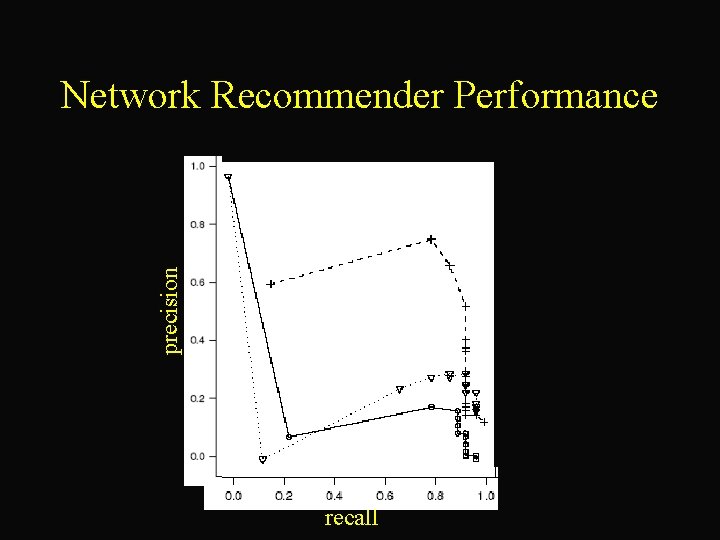
precision Network Recommender Performance recall

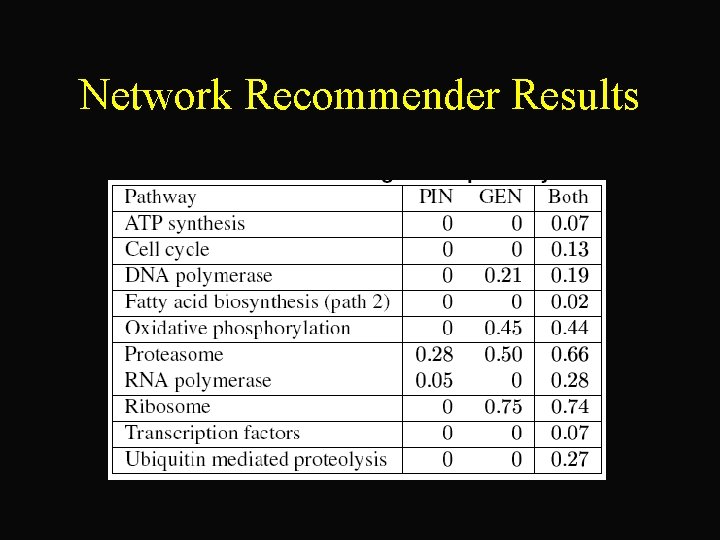
Network Recommender Results

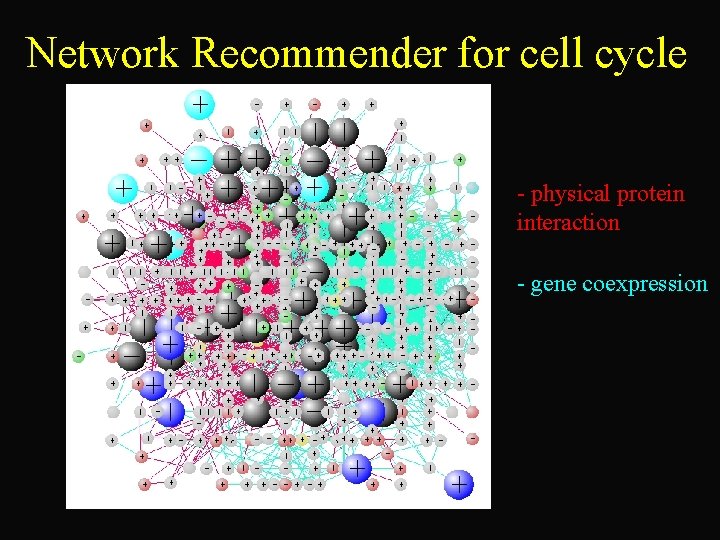
Network Recommender for cell cycle - physical protein interaction - gene coexpression

Supplemental Material

Genetic interactions