Repetitive DNA and nextgeneration sequencing computational challenges and




- Slides: 17

Repetitive DNA and nextgeneration sequencing: computational challenges and solutions TJ Treangen, SL Salzberg Nature Reviews Genetics, 2011 Chen Bichao

Scope 1. Introduction of Repetitive DNA 2. Mapping Assembly 3. De novo Assembly 4. RNA-Seq 5. Conclusions

Introduction of Repetitive DNA

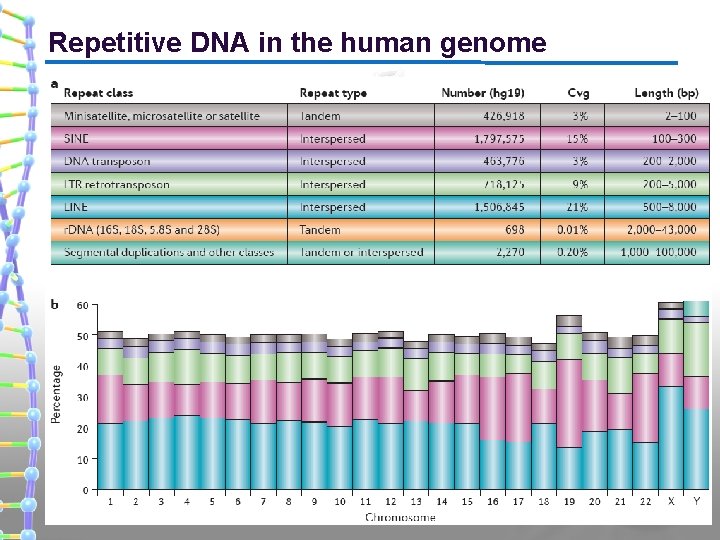
Repetitive DNA in the human genome

Mapping Assembly

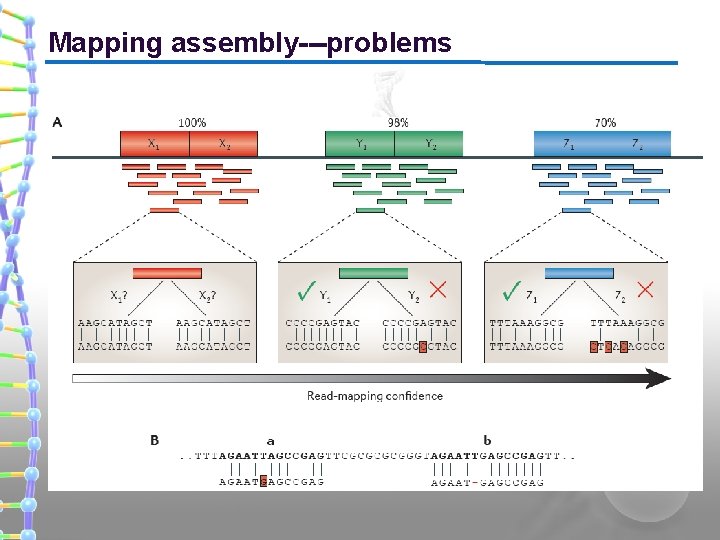
Mapping assembly---problems

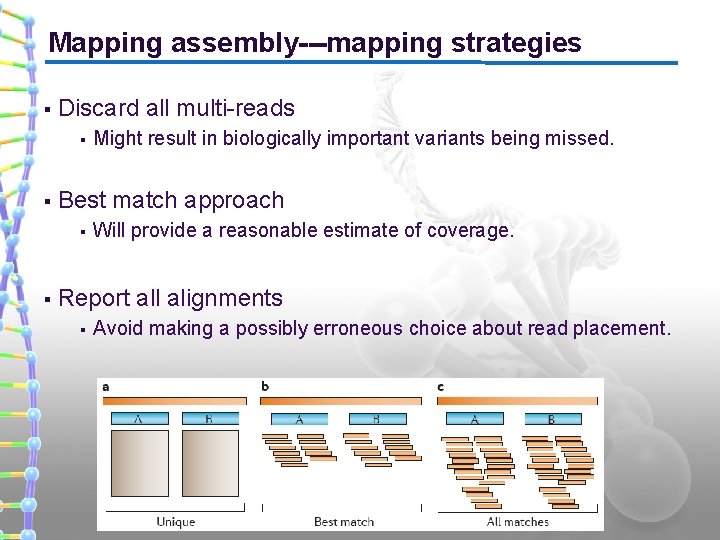
Mapping assembly---mapping strategies § Discard all multi-reads § § Best match approach § § Might result in biologically important variants being missed. Will provide a reasonable estimate of coverage. Report all alignments § Avoid making a possibly erroneous choice about read placement.

De novo Assembly

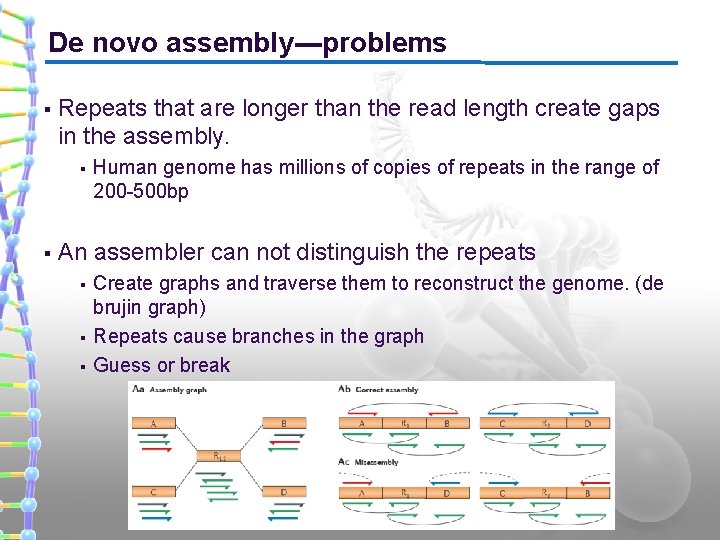
De novo assembly---problems § Repeats that are longer than the read length create gaps in the assembly. § § Human genome has millions of copies of repeats in the range of 200 -500 bp An assembler can not distinguish the repeats § § § Create graphs and traverse them to reconstruct the genome. (de brujin graph) Repeats cause branches in the graph Guess or break

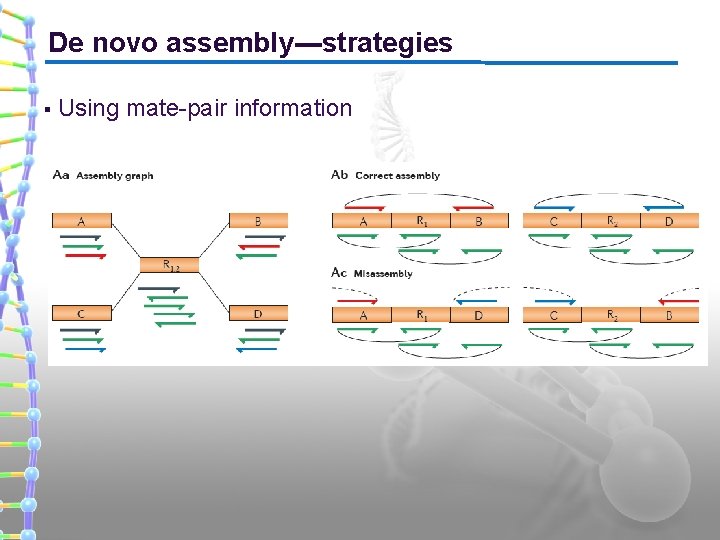
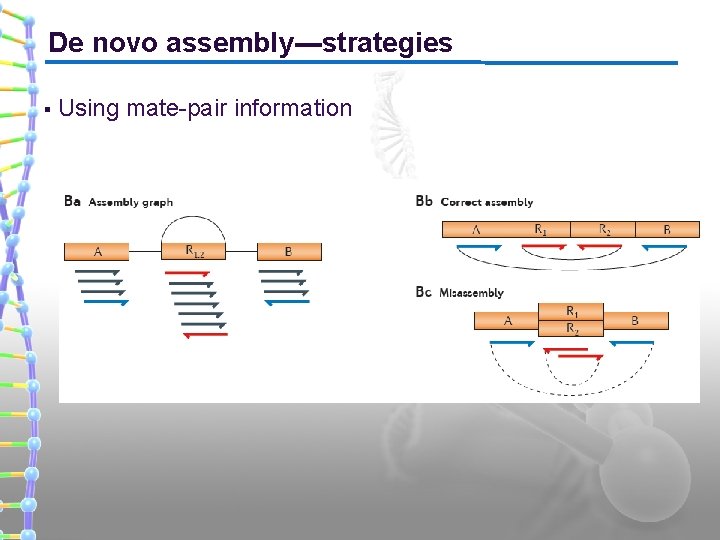
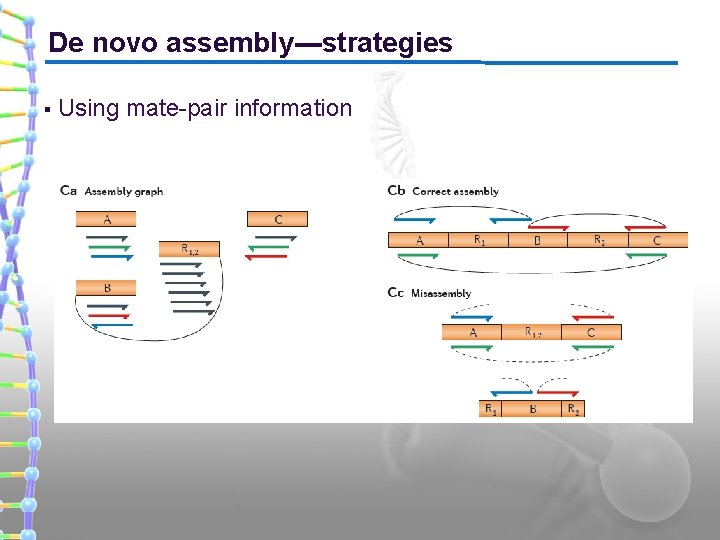
De novo assembly---strategies § Using mate-pair information

De novo assembly---strategies § Using mate-pair information

De novo assembly---strategies § Using mate-pair information

De novo assembly---strategies § Using mate-pair information § Compute statistics on the depth of coverage § § § Assume the genome is uniformly covered Identify the repeats Combination of strategies

RNA-Seq

RNA-seq---problems and strategies § Read splicing § § § Aligning a read to two physically separate locations False positives Strategy for spliced alignment § § Longer sequences align on both sides of each splice site, doesn’t work on fusion genes Exclude any read with more than one (or N) alignment(s) § Estimate gene expression level § Strategy for estimating gene expression § Distribute multi-reads in proportion to the number of reads that map to unique regions of each transcript

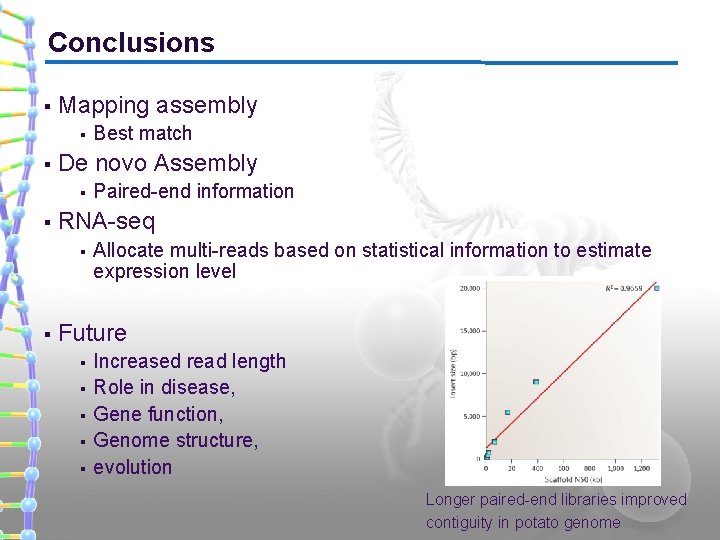
Conclusions § Mapping assembly § § De novo Assembly § § Paired-end information RNA-seq § § Best match Allocate multi-reads based on statistical information to estimate expression level Future § § § Increased read length Role in disease, Gene function, Genome structure, evolution Longer paired-end libraries improved contiguity in potato genome

Thank you. Q&A