Proteomics Technology and Cell Signaling Presenter Ido Tal












- Slides: 49

Proteomics: Technology and Cell Signaling Presenter: Ido Tal Advisor: Prof. Michal Linial 31. 5. 15 י"ג סיון תשע"ה

Outline �Proteomics motivation and background �Identification and quantification technologies – �Antibodies based (e. g. Western Blot) � 2 D Gel �Mass Spectrometry (MS) �SILAC �MS based study �Identified phosphopeptides in signaling cascade �Understanding the function of proteins. �Identified phospho-sites. �difference in phospho-sites �Conclusions and thoughts

What are the functions of proteins? �Replicating DNA �Sensing the environment �Control and Regulation �Enzymes �Transporting molecules �Protecting against pathogens �etc


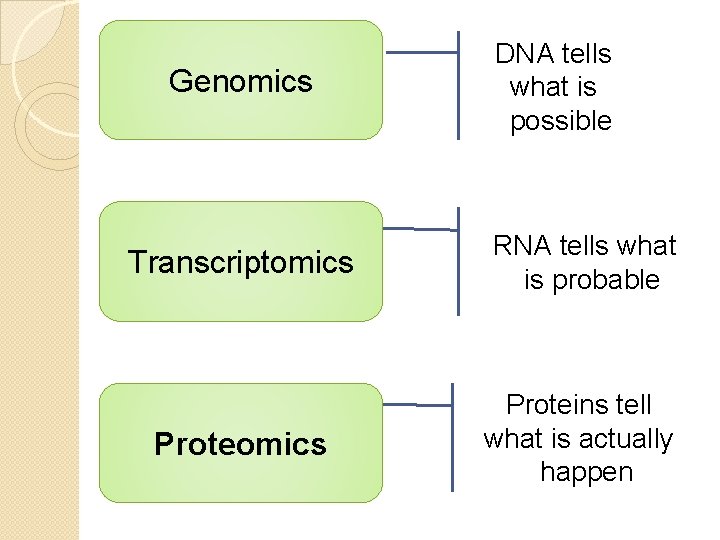
Genomics DNA tells what is possible Transcriptomics RNA tells what is probable Proteomics Proteins tell what is actually happen

Proteome �“The total protein complement able to be encoded by a given genome” “Entire protein complement expressed by a genome, or by a cell or tissue type” Prof. Marc Wilkins, 1995

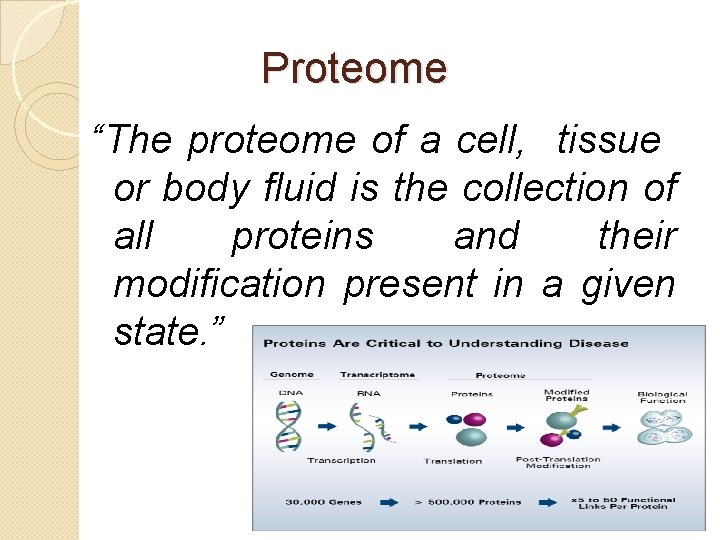
Proteome “The proteome of a cell, tissue or body fluid is the collection of all proteins and their modification present in a given state. ”

What we want to investigate? �The sequence (in amino acids) of a protein �Post translational modification (PTM) �Spatial localization �Identification �Quantity �Protein-protein interactions � 3 D-structure of all proteins �Clinical related (Biomarkers)

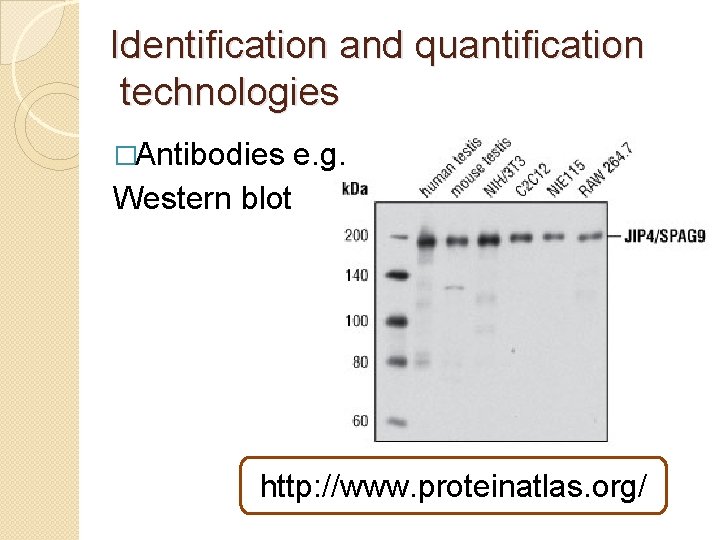
Identification and quantification technologies �Antibodies e. g. Western blot http: //www. proteinatlas. org/

Identification and quantification technologies � 2 D gel

Identification and quantification technologies �Comparative analysis

Identification and quantification technologies �Mass Spectrometry (MS) �SILAC

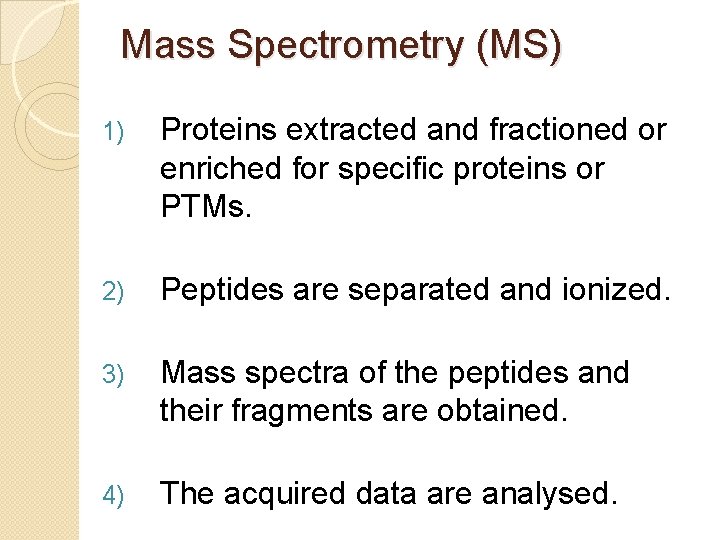
Mass Spectrometry (MS) 1) Proteins extracted and fractioned or enriched for specific proteins or PTMs. 2) Peptides are separated and ionized. 3) Mass spectra of the peptides and their fragments are obtained. 4) The acquired data are analysed.

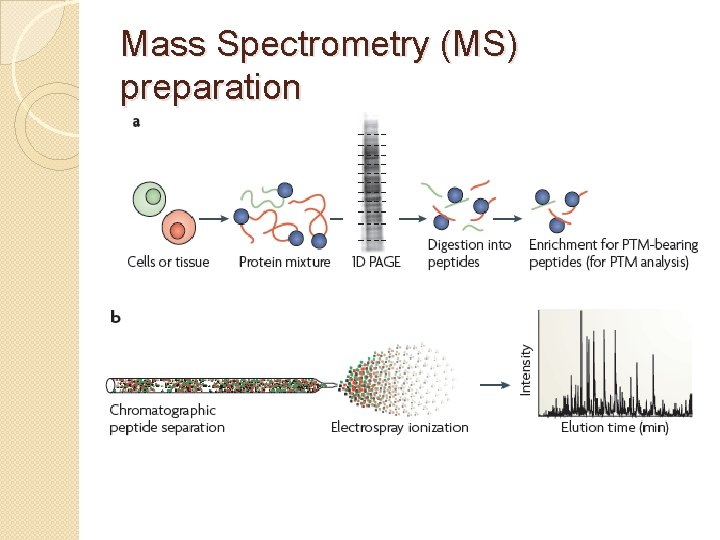
Mass Spectrometry (MS) preparation

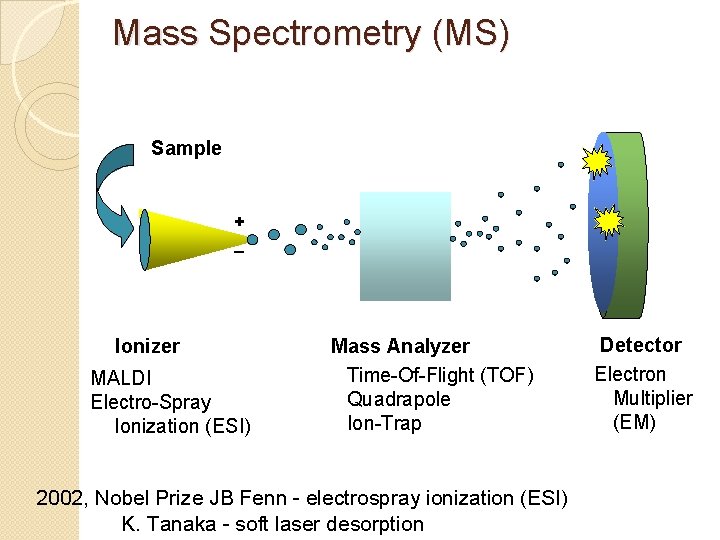
Mass Spectrometry (MS) Sample + _ Ionizer MALDI Electro-Spray Ionization (ESI) Mass Analyzer Time-Of-Flight (TOF) Quadrapole Ion-Trap 2002, Nobel Prize JB Fenn - electrospray ionization (ESI) K. Tanaka - soft laser desorption Detector Electron Multiplier (EM)

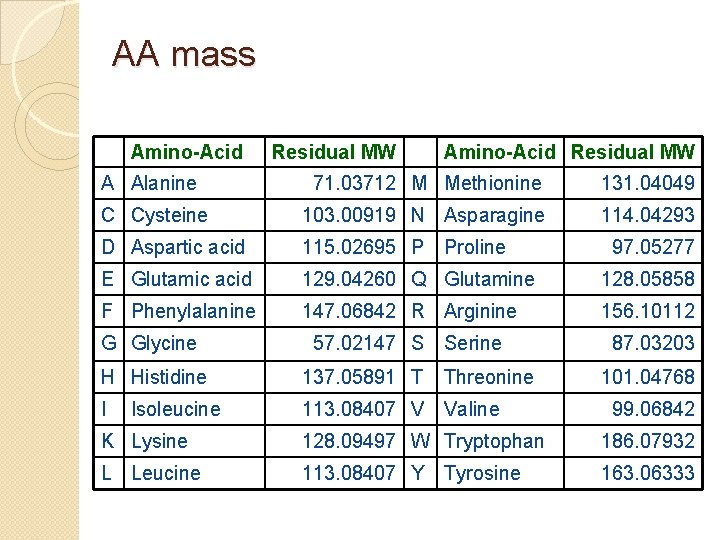
AA mass Amino-Acid Residual MW A Alanine 71. 03712 M Methionine 131. 04049 C Cysteine 103. 00919 N Asparagine 114. 04293 D Aspartic acid 115. 02695 P E Glutamic acid 129. 04260 Q Glutamine 128. 05858 F Phenylalanine 147. 06842 R Arginine 156. 10112 G Glycine 57. 02147 S Proline Serine H Histidine 137. 05891 T Threonine I 113. 08407 V Valine Isoleucine 97. 05277 87. 03203 101. 04768 99. 06842 K Lysine 128. 09497 W Tryptophan 186. 07932 L Leucine 113. 08407 Y 163. 06333 Tyrosine

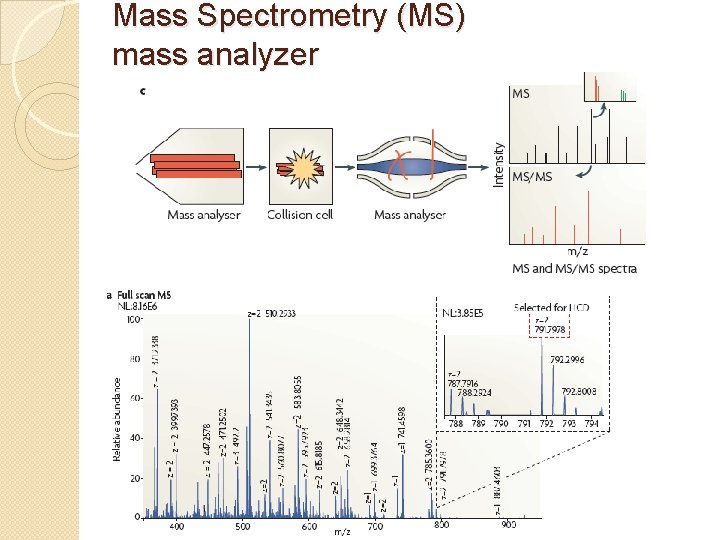
Mass Spectrometry (MS) mass analyzer

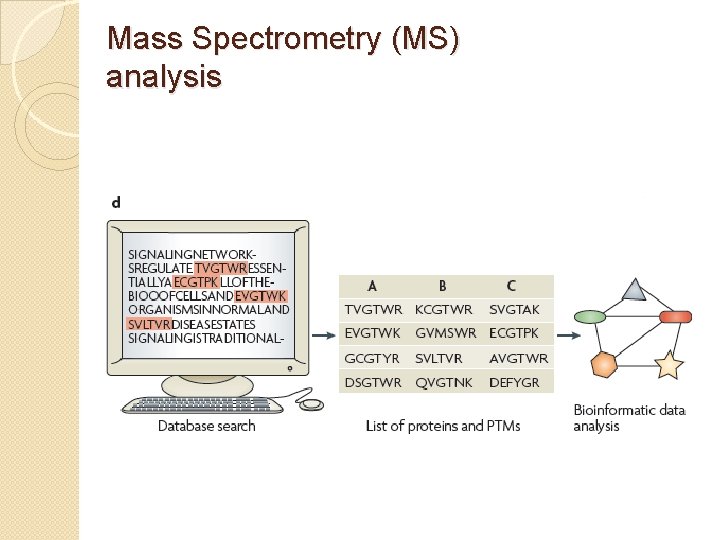
Mass Spectrometry (MS) analysis

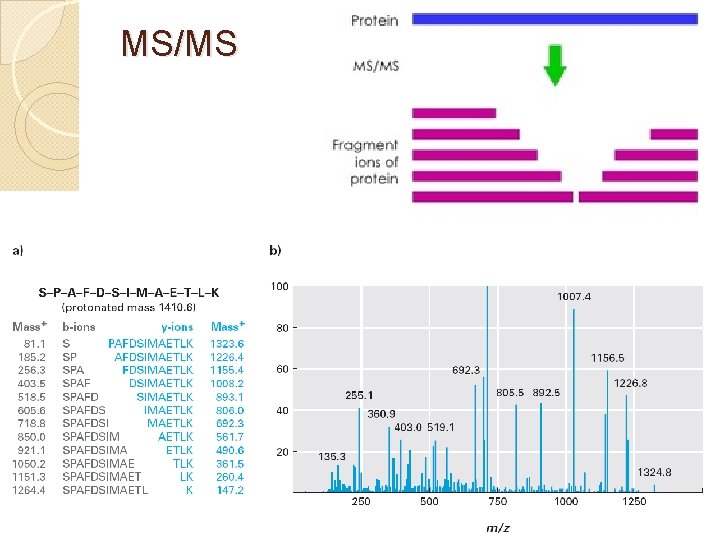
MS/MS

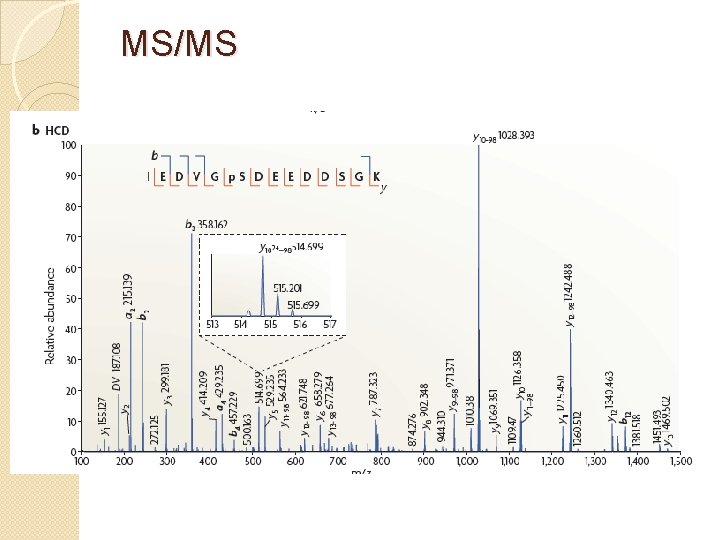
MS/MS

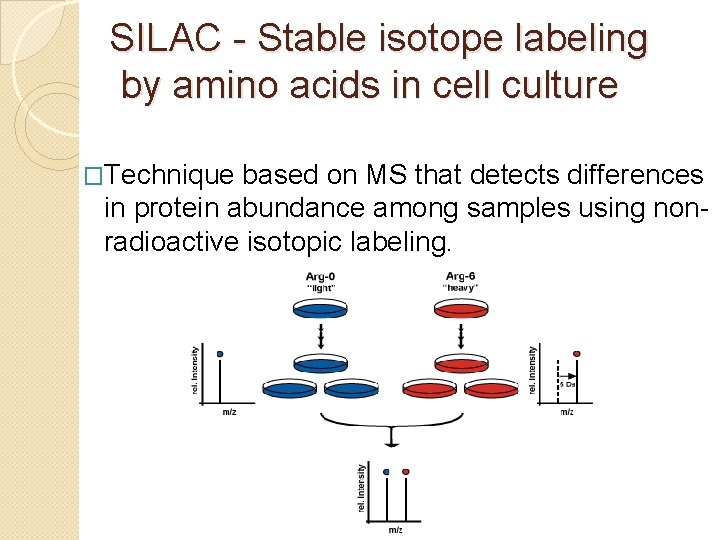
SILAC - Stable isotope labeling by amino acids in cell culture �Technique based on MS that detects differences in protein abundance among samples using nonradioactive isotopic labeling.

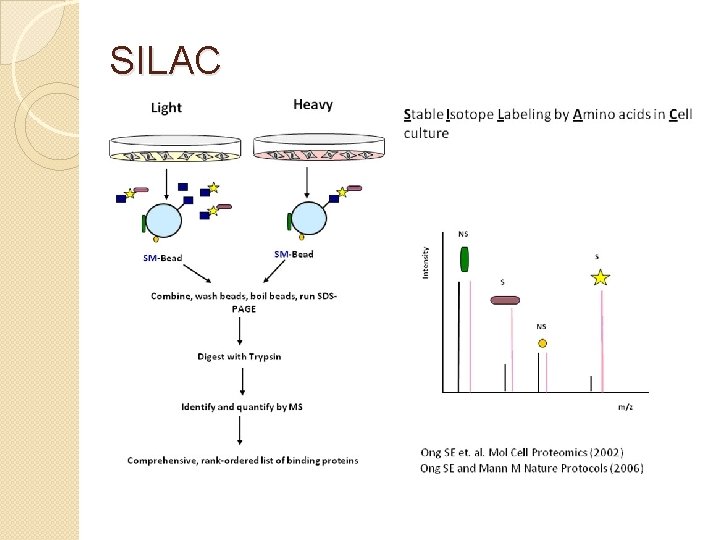
SILAC

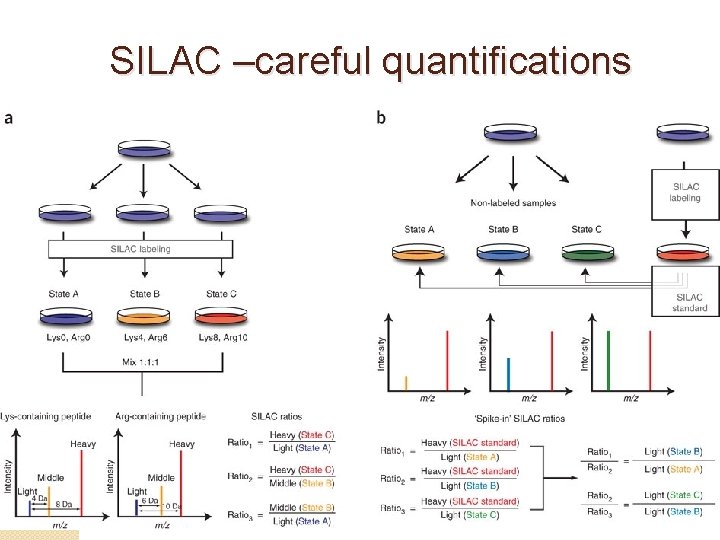
SILAC –careful quantifications

Matthias Mann Education: Physics, Mathematics Chemical Engineering Research Max-Planck-Institute of Biochemistry (Director) Martinsried Germany

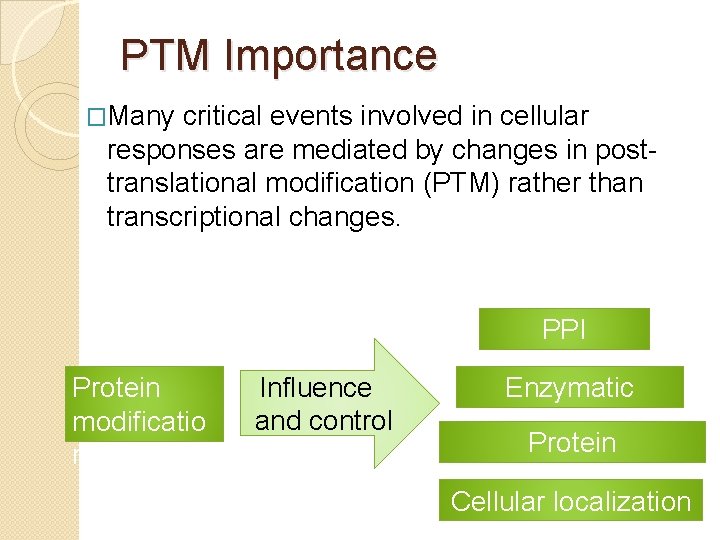
PTM Importance �Many critical events involved in cellular responses are mediated by changes in posttranslational modification (PTM) rather than transcriptional changes. PPI Protein modificatio n Influence and control Enzymatic activity Protein conformation Cellular localization

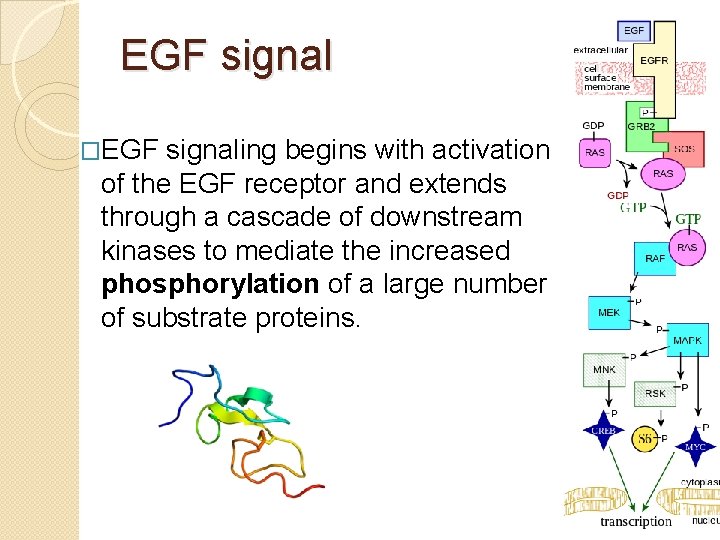
EGF signal �EGF signaling begins with activation of the EGF receptor and extends through a cascade of downstream kinases to mediate the increased phosphorylation of a large number of substrate proteins.

Previous studies �Focus on “early events” �Using anti-phosphotyrosine antibodies, which exclude detection of downstream serine/threonine kinase signaling �large numbers of phosphorylation sites, but without a functional context. �No dynamic view (the sequence of events) �Very few proteins identified in the signaling

The research question Study the global in vivo phosphoproteome and its temporal dynamics upon growth-factor stimulation.



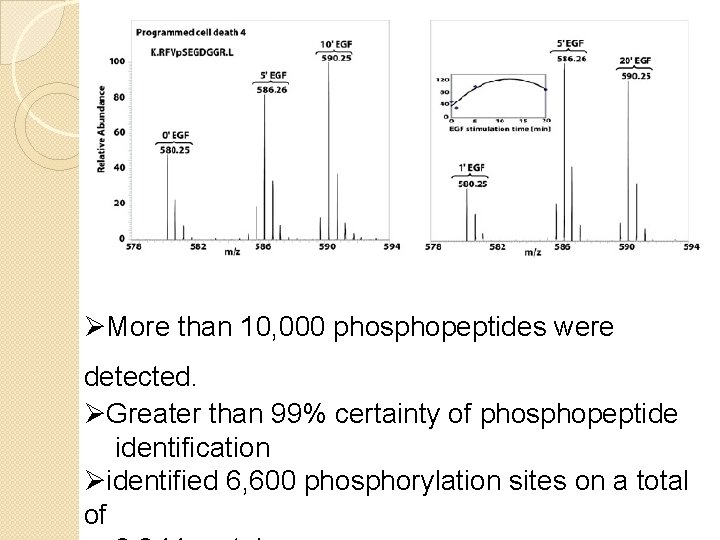
ØMore than 10, 000 phosphopeptides were detected. ØGreater than 99% certainty of phosphopeptide identification Øidentified 6, 600 phosphorylation sites on a total of

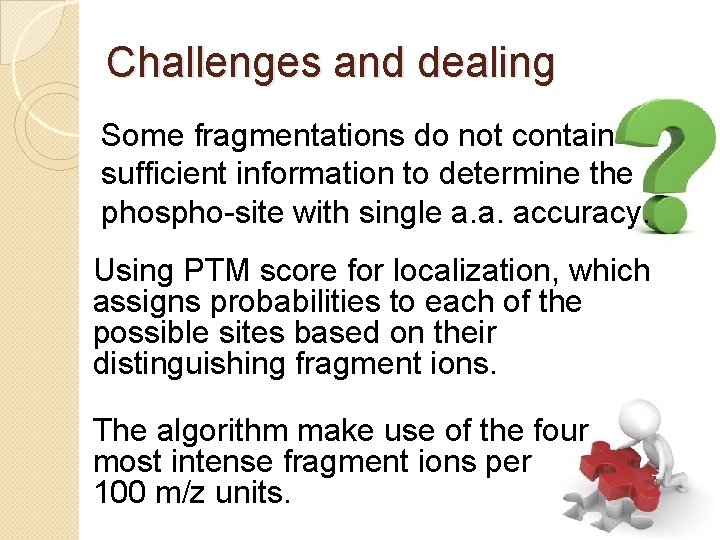
Challenges and dealing Some fragmentations do not contain sufficient information to determine the phospho-site with single a. a. accuracy. Using PTM score for localization, which assigns probabilities to each of the possible sites based on their distinguishing fragment ions. The algorithm make use of the four most intense fragment ions per 100 m/z units.

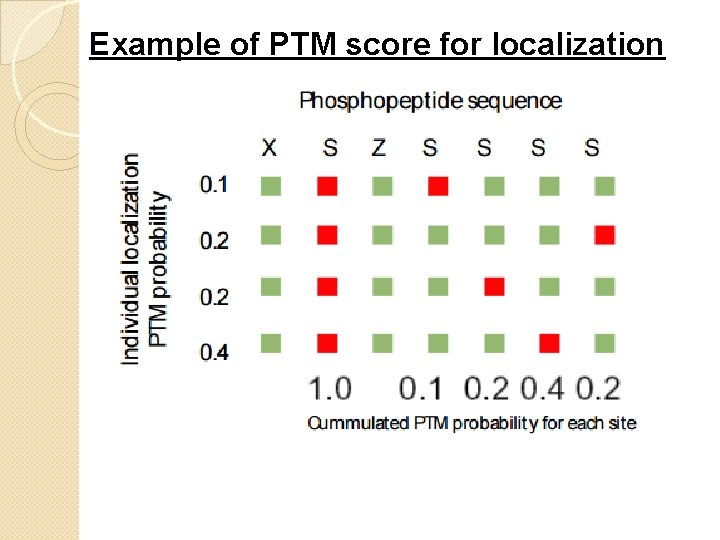
Example of PTM score for localization

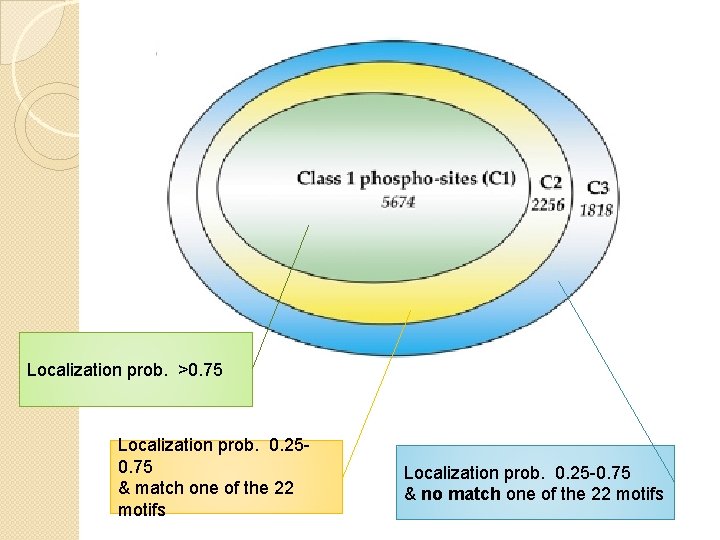
Challenges and dealing In addition take 22 consensus motifs from 15 of the most common kinases and matched all candidate phospho-site against these motifs. Given the peptide sequence and number of phospho-sites, group all peptide into 4 categories depending on the PTM score and motifs.

Localization prob. >0. 75 Localization prob. 0. 250. 75 & match one of the 22 motifs Localization prob. 0. 25 -0. 75 & no match one of the 22 motifs

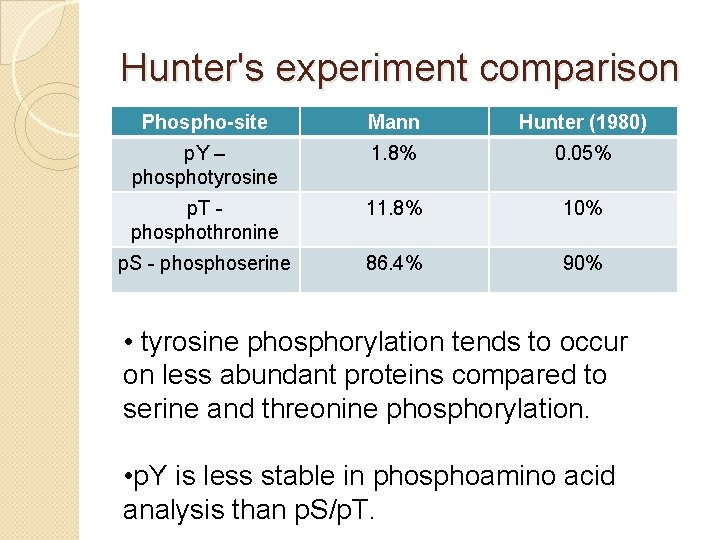
Hunter's experiment comparison Phospho-site Mann Hunter (1980) p. Y – phosphotyrosine 1. 8% 0. 05% p. T phosphothronine 11. 8% 10% p. S - phoserine 86. 4% 90% • tyrosine phosphorylation tends to occur on less abundant proteins compared to serine and threonine phosphorylation. • p. Y is less stable in phosphoamino acid analysis than p. S/p. T.

Database comparison �More than 90% of Mann’s experiment sites were novel with respect to Swiss. Port database. �Close to 90% of Mann’s experiment sites were novel compared with his previous studies.

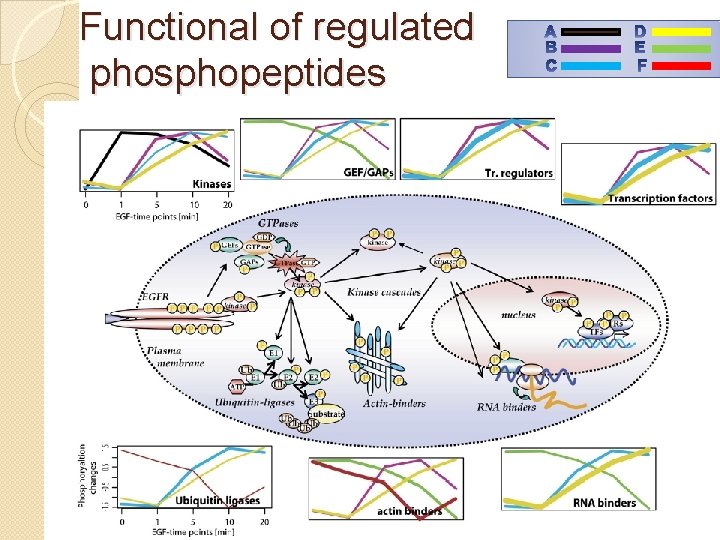
functional context Clustering with Fuzzy c-means – Each profile is assigned a grade of membership for a set of clusters. Found 6 clusters: 4 with upregulated phosphopeptides 2 with downregulated phosphopeptides



Functional of regulated phosphopeptides

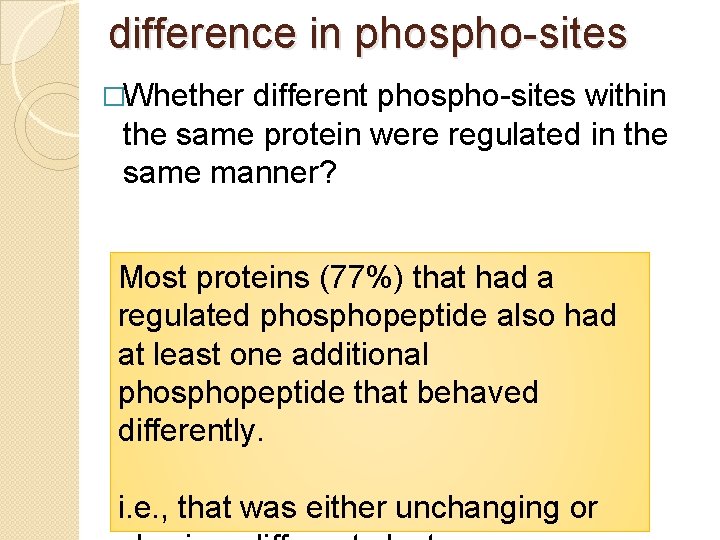
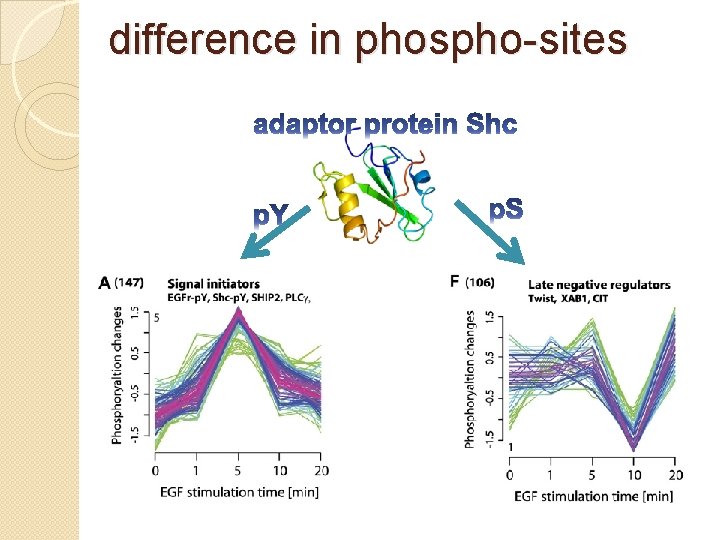
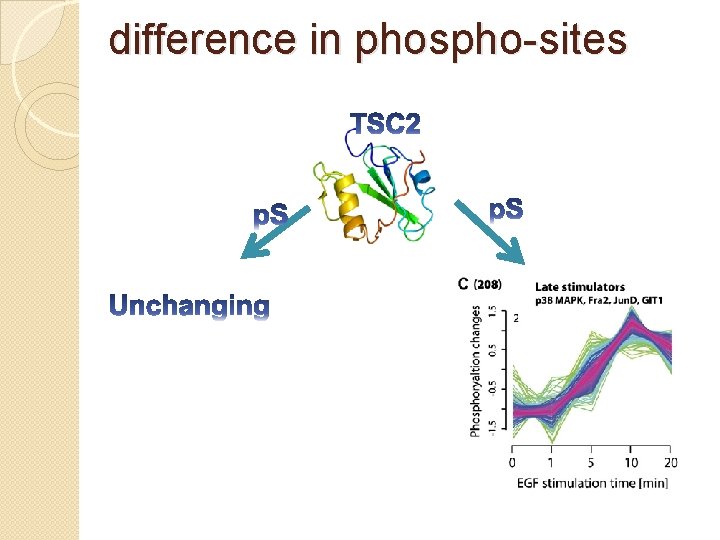
difference in phospho-sites �Whether different phospho-sites within the same protein were regulated in the same manner? Most proteins (77%) that had a regulated phosphopeptide also had at least one additional phosphopeptide that behaved differently. i. e. , that was either unchanging or

difference in phospho-sites

difference in phospho-sites

Difference in phospho-sites �For obtain accurate and functionally relevant understanding of activation kinetics we need to measure site specifically. �Protein phosphorylation typically serves different functions on different sites of the protein.

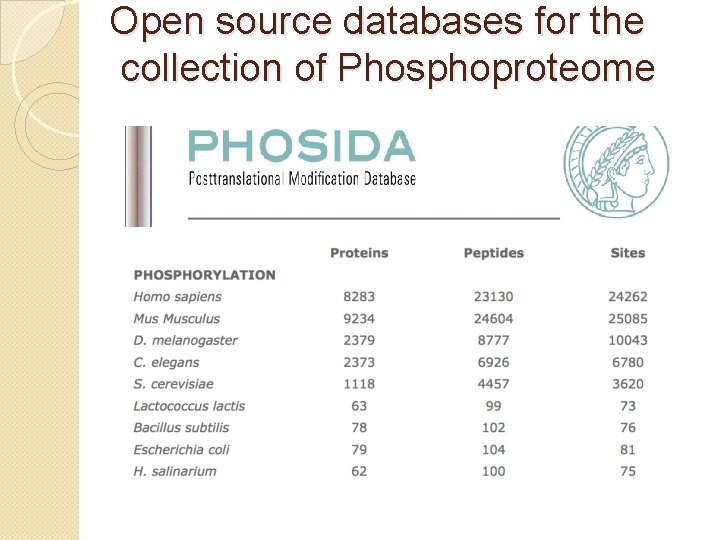
Open source databases for the collection of Phosphoproteome Phosida allows the retrieval of • phosphorylation • acetylation • N-glycosylation of any protein of interest

Open source databases for the collection of Phosphoproteome

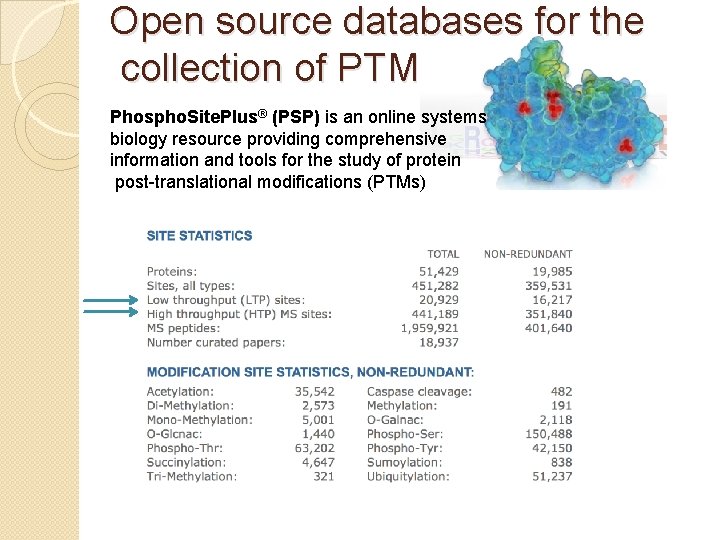
Open source databases for the collection of PTM Phospho. Site. Plus® (PSP) is an online systems biology resource providing comprehensive information and tools for the study of protein post-translational modifications (PTMs)

Conclusions �Importance of proteomics. �Identification and quantification technologies - MS and SILAC and using them for Identified phosphopeptides in signaling cascade �Understanding the function of proteins in signaling using Fuzzy c-means �Identified phospho-sites and the function that can hidden in their difference.