Protein Classification Protein Classification Given a new protein




- Slides: 48

Protein Classification

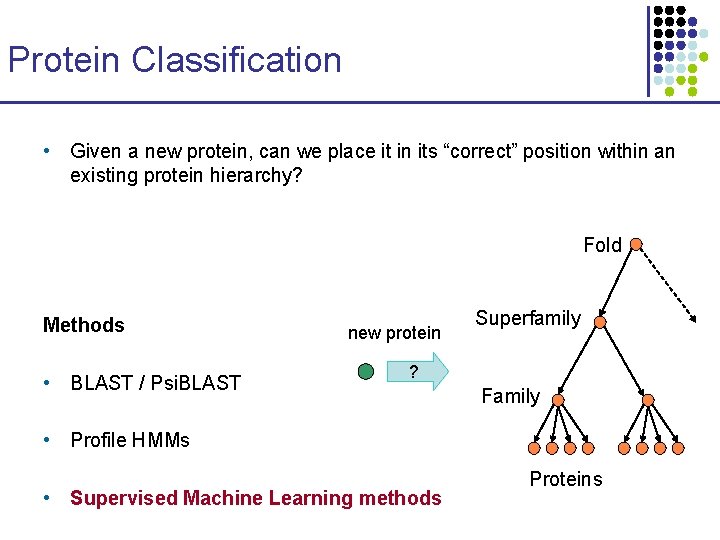
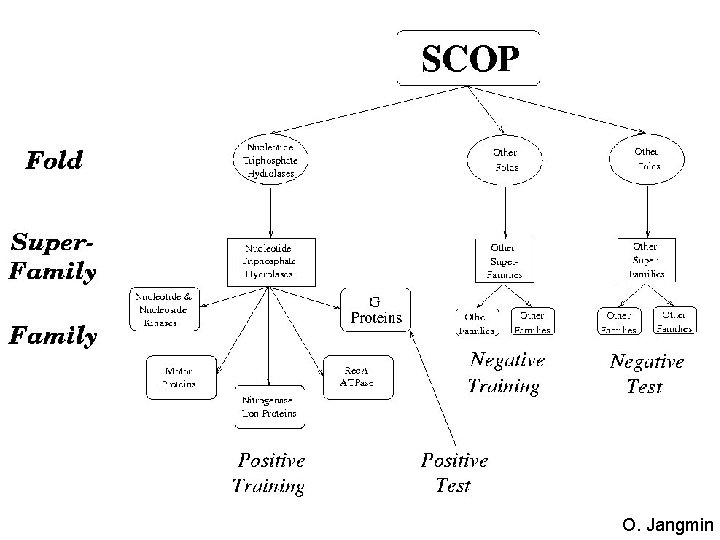
Protein Classification • Given a new protein, can we place it in its “correct” position within an existing protein hierarchy? Fold Methods • BLAST / Psi. BLAST new protein Superfamily ? Family • Profile HMMs • Supervised Machine Learning methods Proteins

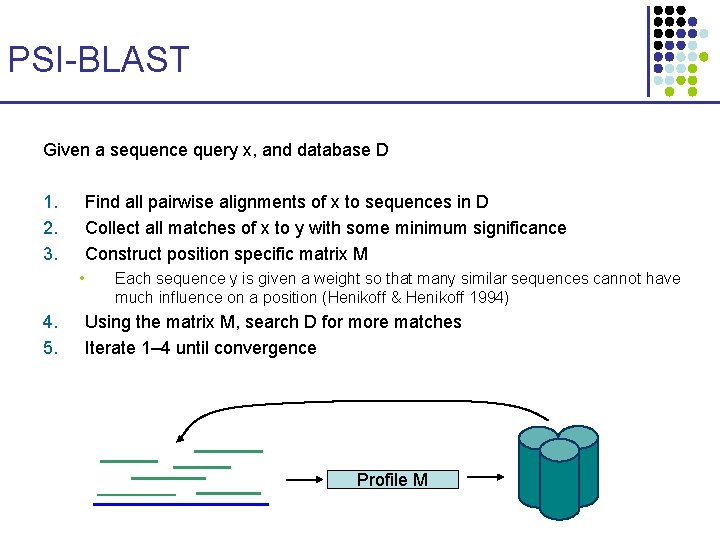
PSI-BLAST Given a sequence query x, and database D 1. 2. 3. Find all pairwise alignments of x to sequences in D Collect all matches of x to y with some minimum significance Construct position specific matrix M • 4. 5. Each sequence y is given a weight so that many similar sequences cannot have much influence on a position (Henikoff & Henikoff 1994) Using the matrix M, search D for more matches Iterate 1– 4 until convergence Profile M

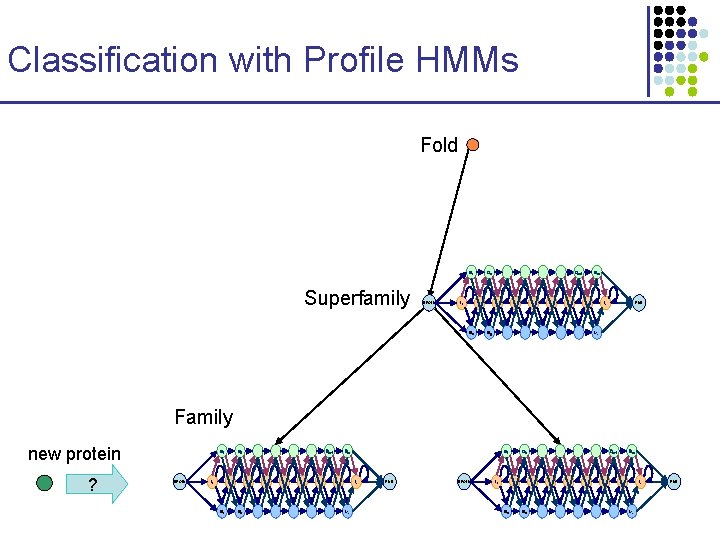
Classification with Profile HMMs Fold D 1 Superfamily BEGIN I 0 Dm-1 D 2 I 1 M 1 Dm Im-1 END Im Mm M 2 Family new protein ? D 1 BEGIN I 0 D 2 I 1 M 1 Dm-1 Dm Im-1 M 2 D 1 Im Mm END BEGIN I 0 D 2 I 1 M 1 Dm-1 Dm Im-1 M 2 Im Mm END

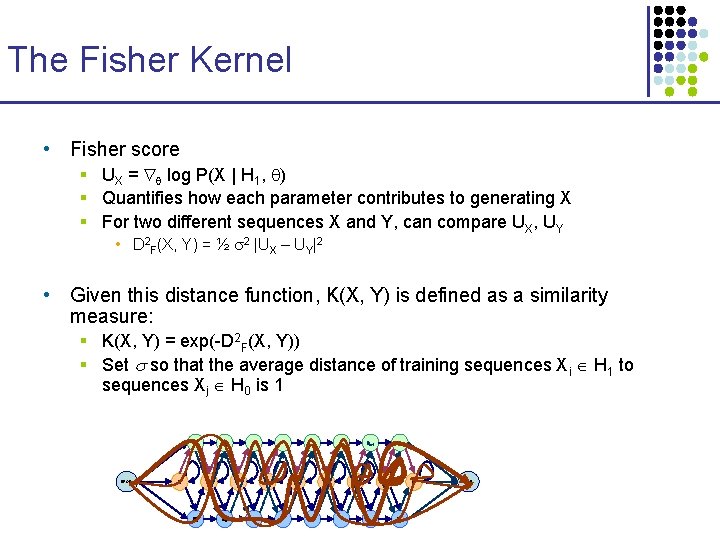
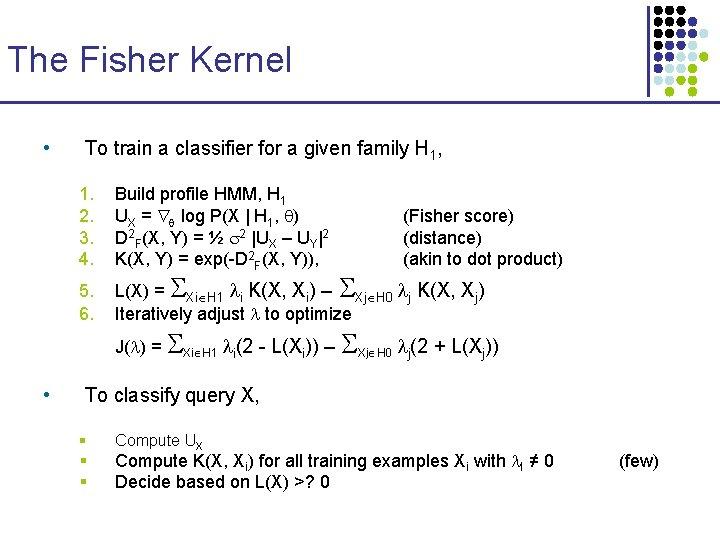
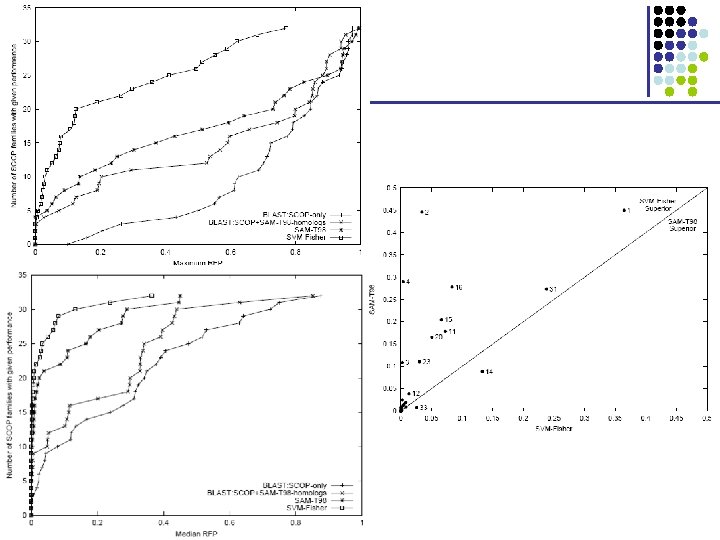
The Fisher Kernel • Fisher score § UX = log P(X | H 1, ) § Quantifies how each parameter contributes to generating X § For two different sequences X and Y, can compare UX, UY • D 2 F(X, Y) = ½ 2 |UX – UY|2 • Given this distance function, K(X, Y) is defined as a similarity measure: § K(X, Y) = exp(-D 2 F(X, Y)) § Set so that the average distance of training sequences Xi H 1 to sequences Xj H 0 is 1 D 1 BEGIN I 0 D 2 I 1 M 1 Dm-1 Dm Im-1 M 2 Im Mm END

The Fisher Kernel • To train a classifier for a given family H 1, 1. 2. 3. 4. 5. 6. Build profile HMM, H 1 UX = log P(X | H 1, ) D 2 F(X, Y) = ½ 2 |UX – UY|2 K(X, Y) = exp(-D 2 F(X, Y)), L(X) = Xi H 1 i K(X, Xi) – Xj H 0 j K(X, Xj) Iteratively adjust to optimize J( ) = Xi H 1 i(2 - L(Xi)) – • (Fisher score) (distance) (akin to dot product) Xj H 0 j(2 + L(Xj)) To classify query X, § § § Compute UX Compute K(X, Xi) for all training examples Xi with I ≠ 0 Decide based on L(X) >? 0 (few)

O. Jangmin


QUESTION Running time of Fisher kernel SVM on query X?

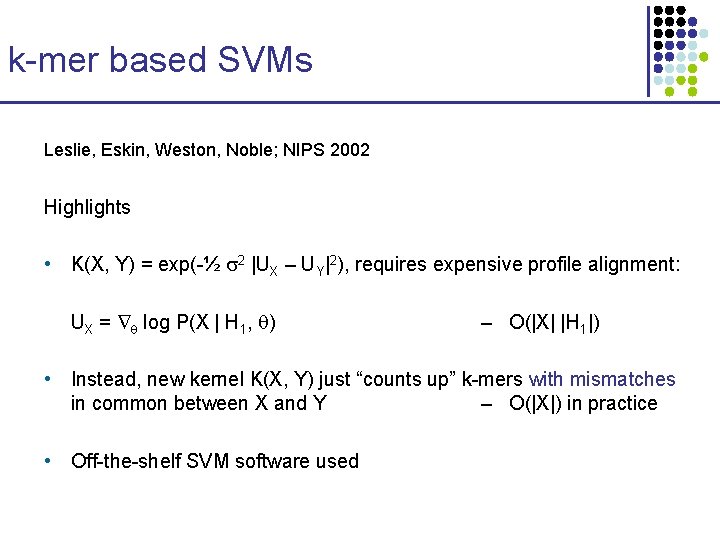
k-mer based SVMs Leslie, Eskin, Weston, Noble; NIPS 2002 Highlights • K(X, Y) = exp(-½ 2 |UX – UY|2), requires expensive profile alignment: UX = log P(X | H 1, ) – O(|X| |H 1|) • Instead, new kernel K(X, Y) just “counts up” k-mers with mismatches in common between X and Y – O(|X|) in practice • Off-the-shelf SVM software used

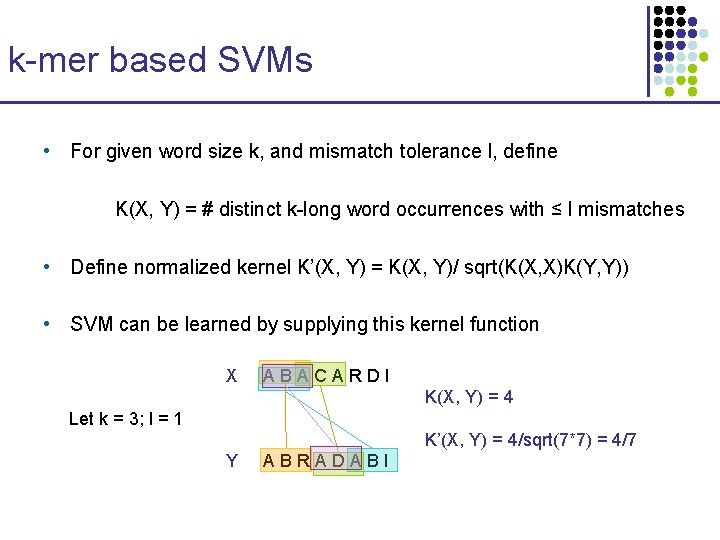
k-mer based SVMs • For given word size k, and mismatch tolerance l, define K(X, Y) = # distinct k-long word occurrences with ≤ l mismatches • Define normalized kernel K’(X, Y) = K(X, Y)/ sqrt(K(X, X)K(Y, Y)) • SVM can be learned by supplying this kernel function X ABACARDI K(X, Y) = 4 Let k = 3; l = 1 K’(X, Y) = 4/sqrt(7*7) = 4/7 Y ABRADABI

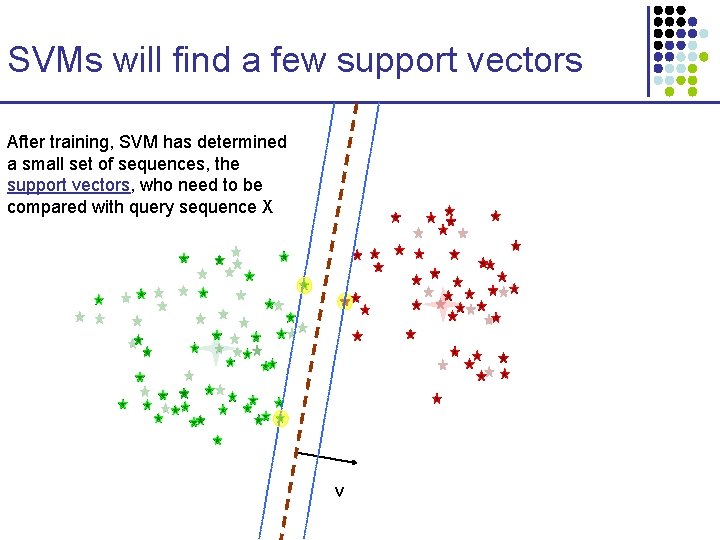
SVMs will find a few support vectors After training, SVM has determined a small set of sequences, the support vectors, who need to be compared with query sequence X v

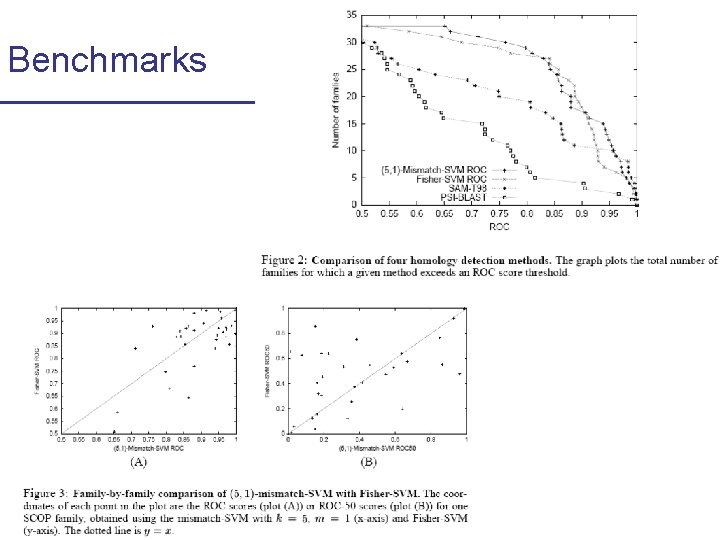
Benchmarks

Semi-Supervised Methods GENERATIVE SUPERVISED METHODS

Semi-Supervised Methods DISCRIMINATIVE SUPERVISED METHODS

Semi-Supervised Methods UNSUPERVISED METHODS Mixture of Centers Data generated by a fixed set of centers (how many? )

Semi-Supervised Methods UNSUPERVISED METHODS Mixture of Centers Data generated by a fixed set of centers (how many? )

Semi-Supervised Methods UNSUPERVISED METHODS Mixture of Centers Data generated by a fixed set of centers (how many? )

Semi-Supervised Methods UNSUPERVISED METHODS Mixture of Centers Data generated by a fixed set of centers (how many? )

Semi-Supervised Methods UNSUPERVISED METHODS Mixture of Centers Data generated by a fixed set of centers (how many? )

Semi-Supervised Methods UNSUPERVISED METHODS Mixture of Centers Data generated by a fixed set of centers (how many? )

Semi-Supervised Methods UNSUPERVISED METHODS Mixture of Centers Data generated by a fixed set of centers (how many? )

Semi-Supervised Methods UNSUPERVISED METHODS Mixture of Centers Data generated by a fixed set of centers (how many? )

Semi-Supervised Methods UNSUPERVISED METHODS Mixture of Centers Data generated by a fixed set of centers (how many? )

Semi-Supervised Methods UNSUPERVISED METHODS Mixture of Centers Data generated by a fixed set of centers (how many? )

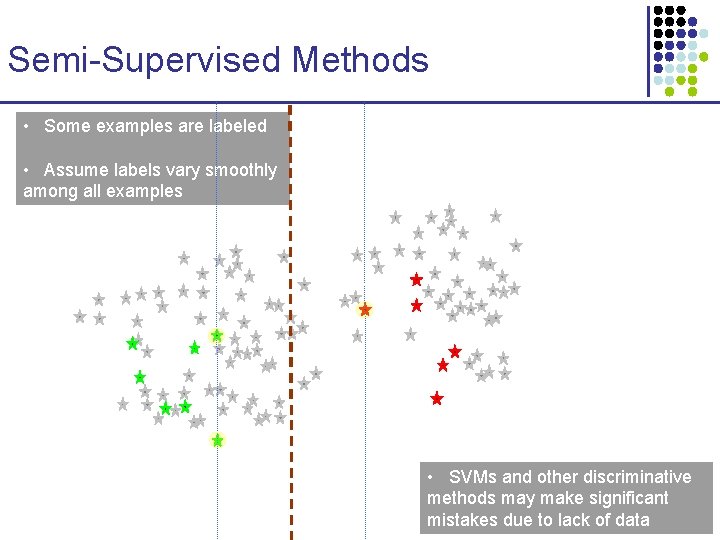
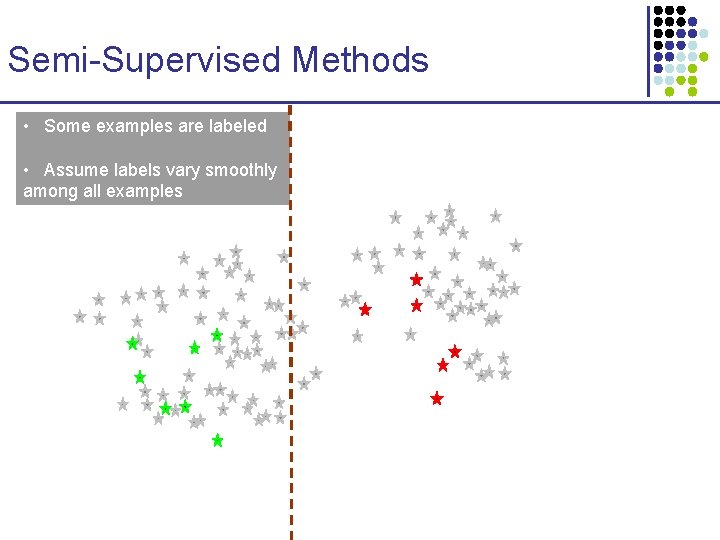
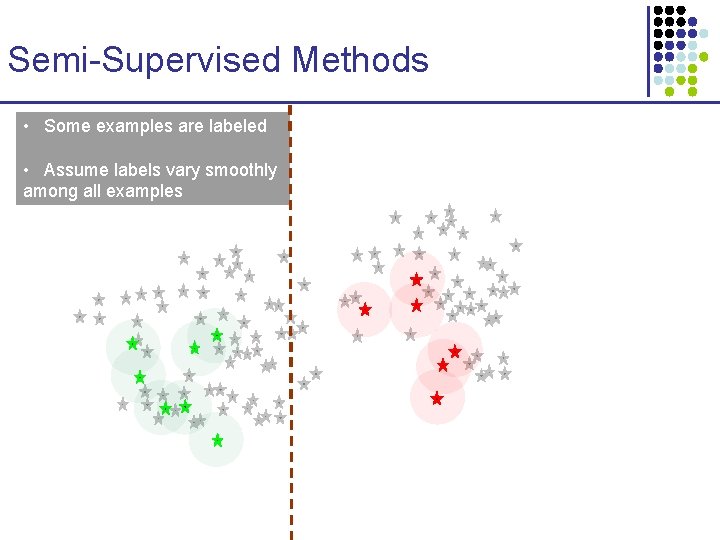
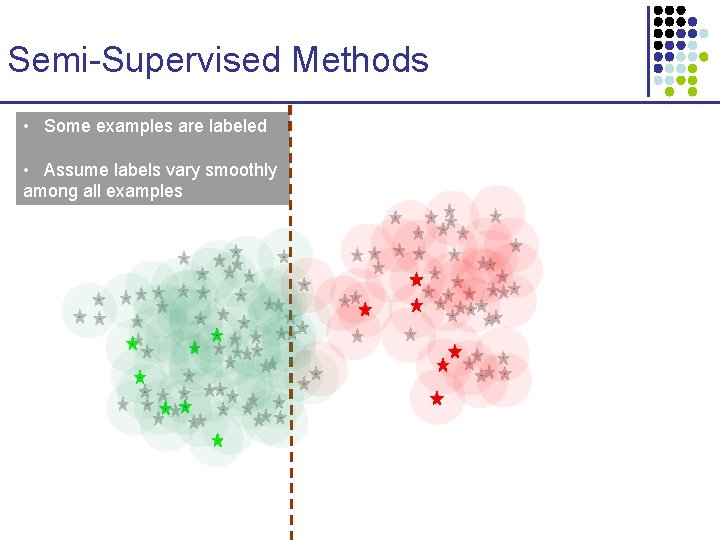
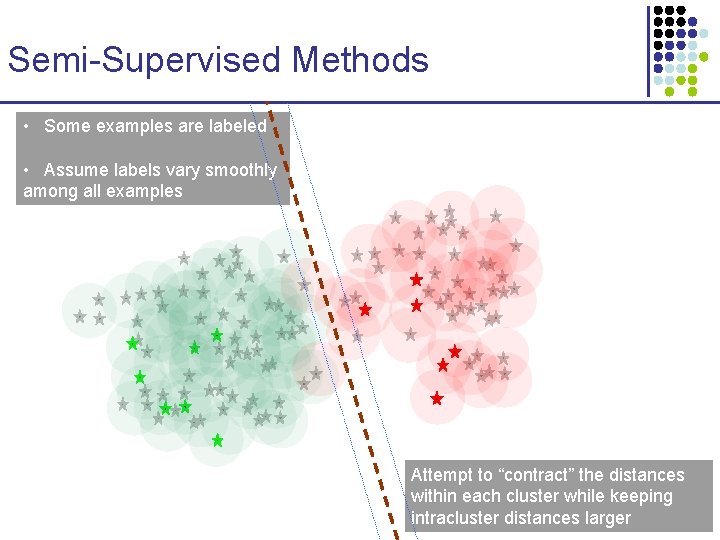
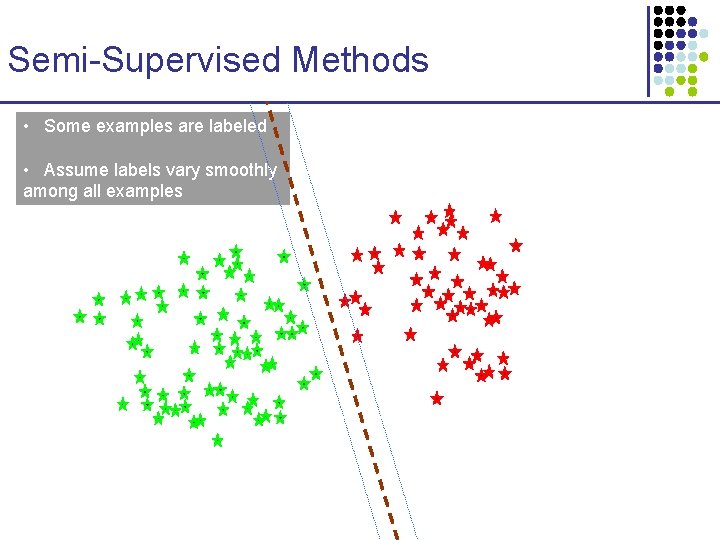
Semi-Supervised Methods • Some examples are labeled • Assume labels vary smoothly among all examples

Semi-Supervised Methods • Some examples are labeled • Assume labels vary smoothly among all examples • SVMs and other discriminative methods may make significant mistakes due to lack of data

Semi-Supervised Methods • Some examples are labeled • Assume labels vary smoothly among all examples

Semi-Supervised Methods • Some examples are labeled • Assume labels vary smoothly among all examples

Semi-Supervised Methods • Some examples are labeled • Assume labels vary smoothly among all examples

Semi-Supervised Methods • Some examples are labeled • Assume labels vary smoothly among all examples Attempt to “contract” the distances within each cluster while keeping intracluster distances larger

Semi-Supervised Methods • Some examples are labeled • Assume labels vary smoothly among all examples

Semi-Supervised Methods 1. Kuang, Ie, Wang, Siddiqi, Freund, Leslie 2005 § 2. A Psi-BLAST profile—based method Weston, Leslie, Elisseeff, Noble, NIPS 2003 § Cluster kernels

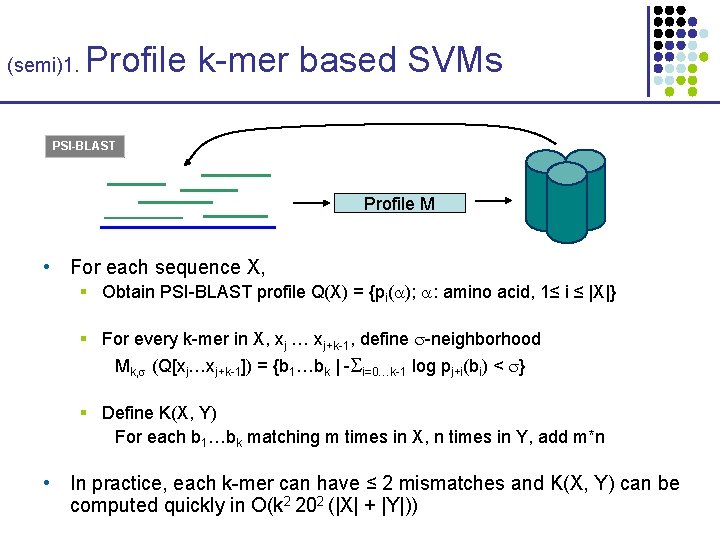
(semi)1. Profile k-mer based SVMs PSI-BLAST Profile M • For each sequence X, § Obtain PSI-BLAST profile Q(X) = {pi( ); : amino acid, 1≤ i ≤ |X|} § For every k-mer in X, xj … xj+k-1, define -neighborhood Mk, (Q[xj…xj+k-1]) = {b 1…bk | - i=0…k-1 log pj+i(bi) < } § Define K(X, Y) For each b 1…bk matching m times in X, n times in Y, add m*n • In practice, each k-mer can have ≤ 2 mismatches and K(X, Y) can be computed quickly in O(k 2 202 (|X| + |Y|))

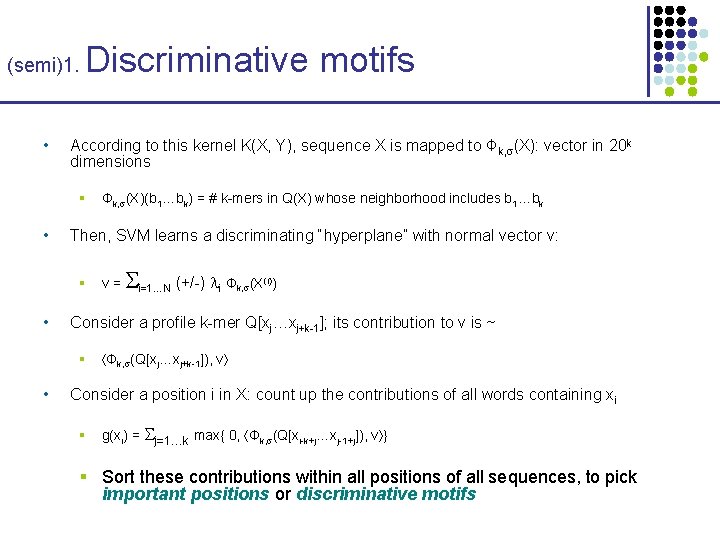
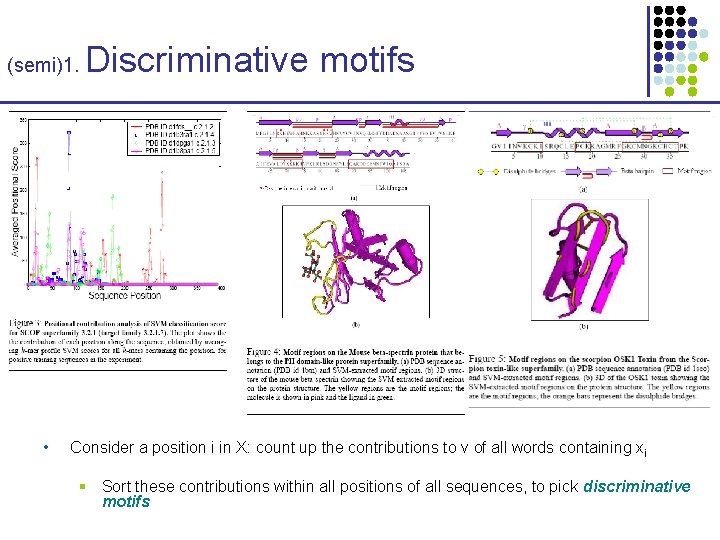
(semi)1. • Discriminative motifs According to this kernel K(X, Y), sequence X is mapped to Φk, (X): vector in 20 k dimensions § • Then, SVM learns a discriminating “hyperplane” with normal vector v: § • v= i=1…N (+/-) i Φ k, (X (i)) Consider a profile k-mer Q[xj…xj+k-1]; its contribution to v is ~ § • Φk, (X)(b 1…bk) = # k-mers in Q(X) whose neighborhood includes b 1…bk Φk, (Q[xj…xj+k-1]), v Consider a position i in X: count up the contributions of all words containing xi § g(xi) = j=1…k max{ 0, Φk, (Q[xi-k+j…xj-1+j]), v } § Sort these contributions within all positions of all sequences, to pick important positions or discriminative motifs

(semi)1. • Discriminative motifs Consider a position i in X: count up the contributions to v of all words containing xi § Sort these contributions within all positions of all sequences, to pick discriminative motifs

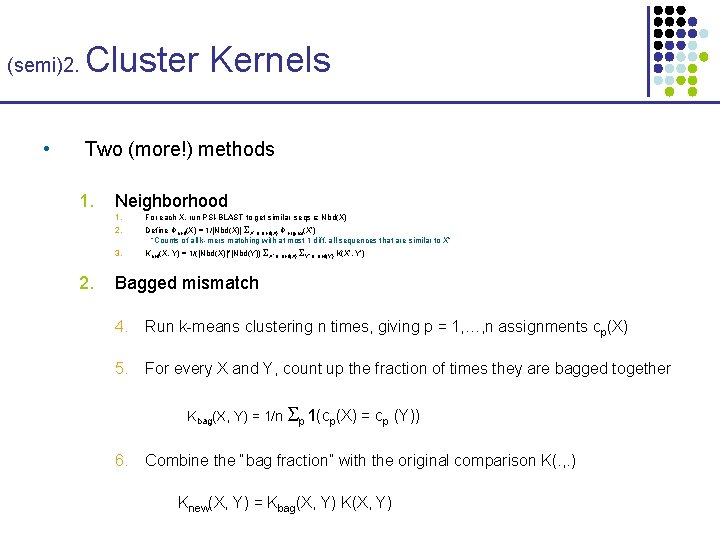
(semi)2. • Cluster Kernels Two (more!) methods 1. Neighborhood 1. For each X, run PSI-BLAST to get similar seqs Nbd(X) 2. Define Φnbd(X) = 1/|Nbd(X)| X’ Nbd(X) Φoriginal(X’) “Counts of all k-mers matching with at most 1 diff. all sequences that are similar to X” 3. Knbd(X, Y) = 1/(|Nbd(X)|*|Nbd(Y)) X’ Nbd(X) Y’ Nbd(Y) K(X’, Y’) 2. Bagged mismatch

(semi)2. • Cluster Kernels Two (more!) methods 1. 2. Neighborhood 1. For each X, run PSI-BLAST to get similar seqs Nbd(X) 2. Define Φnbd(X) = 1/|Nbd(X)| X’ Nbd(X) Φoriginal(X’) “Counts of all k-mers matching with at most 1 diff. all sequences that are similar to X” 3. Knbd(X, Y) = 1/(|Nbd(X)|*|Nbd(Y)) X’ Nbd(X) Y’ Nbd(Y) K(X’, Y’) Bagged mismatch 4. Run k-means clustering n times, giving p = 1, …, n assignments cp(X) 5. For every X and Y, count up the fraction of times they are bagged together Kbag(X, Y) = 1/n p 1(cp(X) = cp (Y)) 6. Combine the “bag fraction” with the original comparison K(. , . ) Knew(X, Y) = Kbag(X, Y) K(X, Y)

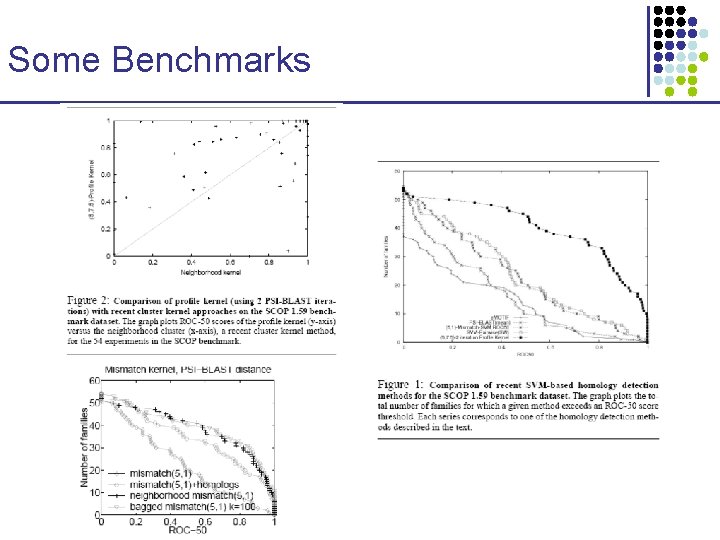
Some Benchmarks

Google-like homology search • The internet and the network of protein homologies have some similarity—scale free • Given query X, Google ranks webpages by a flow algorithm § From each webpage W, linked nbrs receive flow § At time t+1, W sends to nbrs flow it received at time t § Finite, ergodic, aperiodic Markov Chain § Can find stationary distribution efficiently as left eigenvector with eigenvalue 1 • Start with arbitrary probability distribution, and multiply by the transition matrix

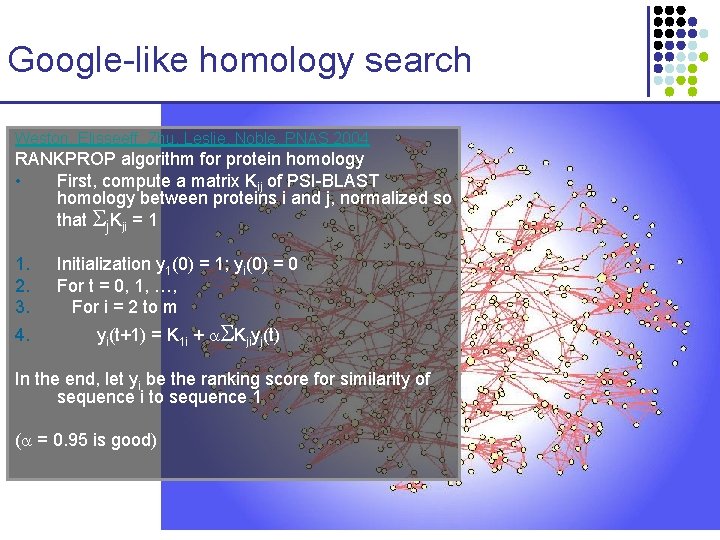
Google-like homology search Weston, Elisseeff, Zhu, Leslie, Noble, PNAS 2004 RANKPROP algorithm for protein homology • First, compute a matrix Kij of PSI-BLAST homology between proteins i and j, normalized so that j. Kji = 1 1. 2. 3. 4. Initialization y 1(0) = 1; yi(0) = 0 For t = 0, 1, …, For i = 2 to m yi(t+1) = K 1 i + Kjiyj(t) In the end, let yi be the ranking score for similarity of sequence i to sequence 1 ( = 0. 95 is good)

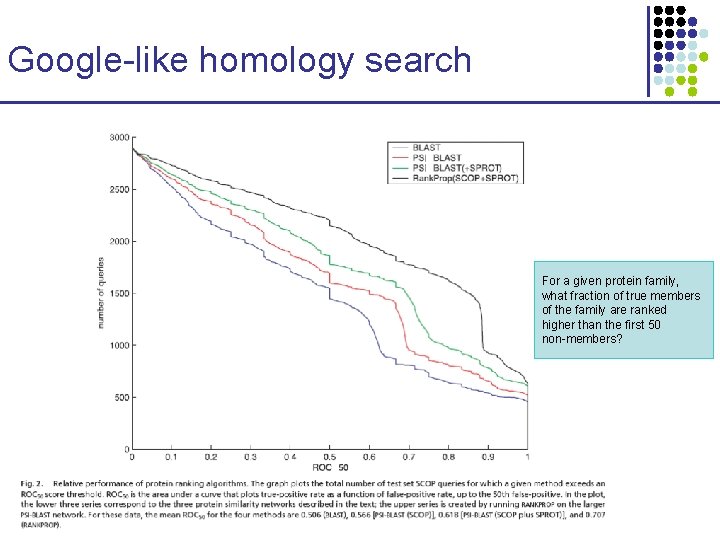
Google-like homology search For a given protein family, what fraction of true members of the family are ranked higher than the first 50 non-members?

Protein Structure Prediction

Protein Structure Determination • Experimental § X-ray crystallography § NMR spectrometry • Computational – Structure Prediction (The Holy Grail) Sequence implies structure, therefore in principle we can predict the structure from the sequence alone

Protein Structure Prediction • ab initio § Use just first principles: energy, geometry, and kinematics • Homology § Find the best match to a database of sequences with known 3 Dstructure • Threading • Meta-servers and other methods

Ab initio Prediction • Sampling the global conformation space § Lattice models / Discrete-state models § Molecular Dynamics • Picking native conformations with an energy function § Solvation model: how protein interacts with water § Pair interactions between amino acids • Predicting secondary structure § Local homology § Fragment libraries

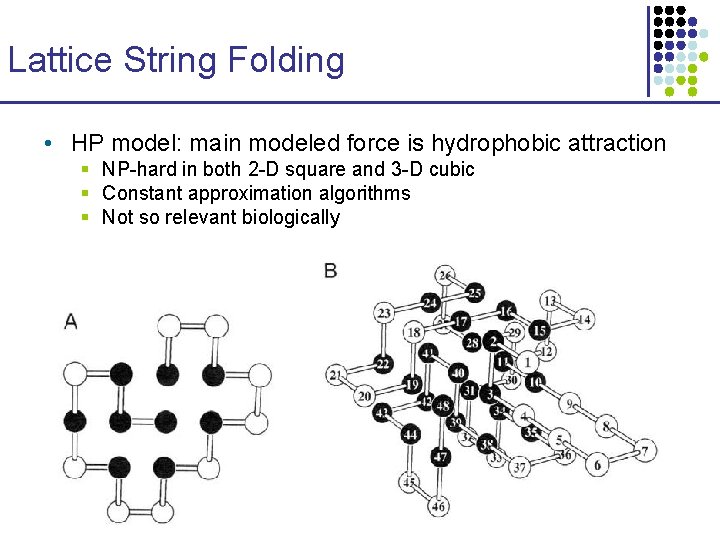
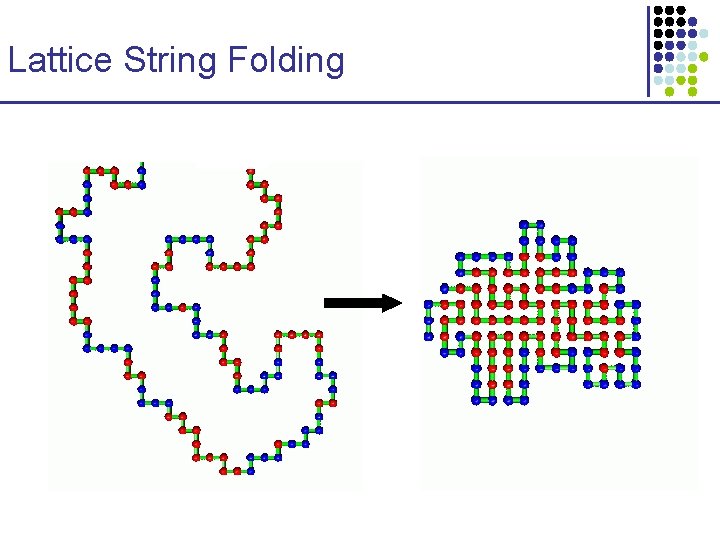
Lattice String Folding • HP model: main modeled force is hydrophobic attraction § NP-hard in both 2 -D square and 3 -D cubic § Constant approximation algorithms § Not so relevant biologically

Lattice String Folding