Predicting Recurrence in Clear Cell Renal Cell Carcinoma





- Slides: 16

Predicting Recurrence in Clear Cell Renal Cell Carcinoma Analysis of TCGA data using Outlier Analysis and GMLVQ Gargi Mukherjee Kevin Raines Srikanth Sastry Sebastian Doniach Gyan Bhanot Michael Biehl … … … Rutgers University, New Jersey Stanford University, California JNC, Bengaluru, India Stanford University, California Rutgers University, New Jersey University of Groningen, The Netherlands 1

overview gene expression in tumor cells specific example: clear cell Renal Cell Carcinomas (cc. RCC) clinical data: recurrence free intervals • outlier analysis: identification of a panel of prognostic genes with respect to recurrence • risk score: prediction of individual recurrence risk based on outlier status w. r. t. selected genes • machine learning: analysis of extreme cases of low / high risk distance based classification and relevance learning (Generalized Matrix Relevance LVQ) WCCI 2016, Vancouver / BC 2 /15

data clear cell Renal Cell Carcinoma (cc. RCC) publicly available datasets: The Cancer Genome Atlas (TCGA) cancergenome. nih. gov also hosted at Broad Institute gdac. broadinstitute. org WCCI 2016, Vancouver / BC 3 /15

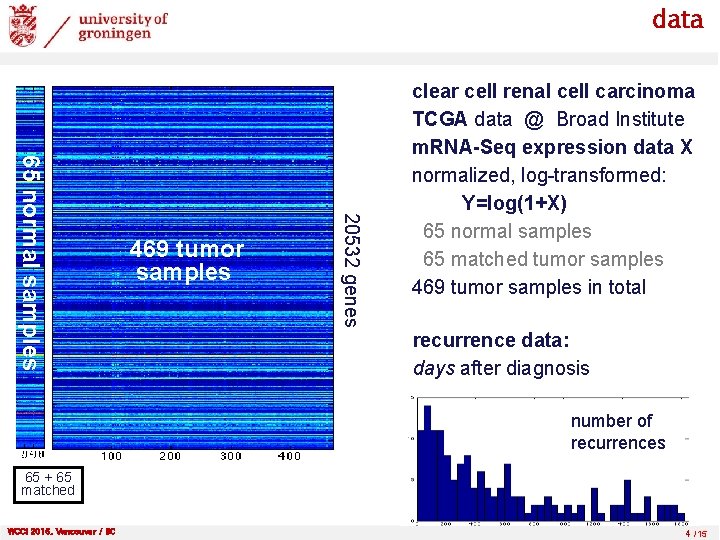
data 20532 genes 65 normal samples 469 tumor samples clear cell renal cell carcinoma TCGA data @ Broad Institute m. RNA-Seq expression data X normalized, log-transformed: Y=log(1+X) 65 normal samples 65 matched tumor samples 469 tumor samples in total recurrence data: days after diagnosis number of recurrences 65 + 65 matched WCCI 2016, Vancouver / BC 4 /15

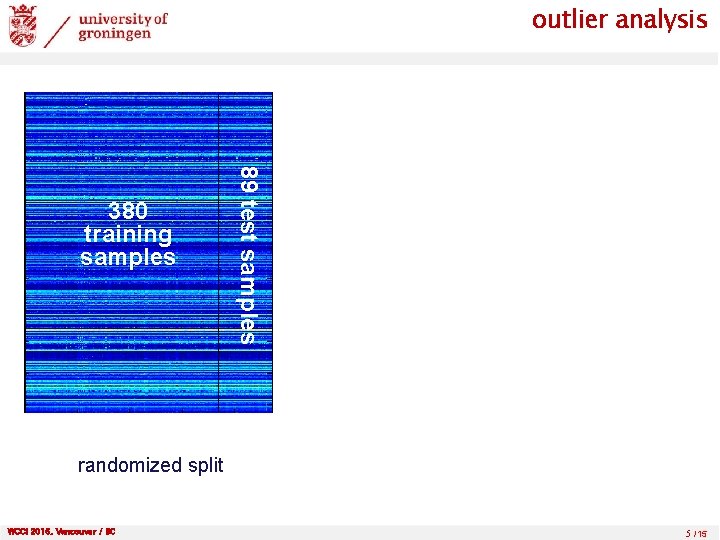
outlier analysis 89 test samples 380 training samples randomized split WCCI 2016, Vancouver / BC 5 /15

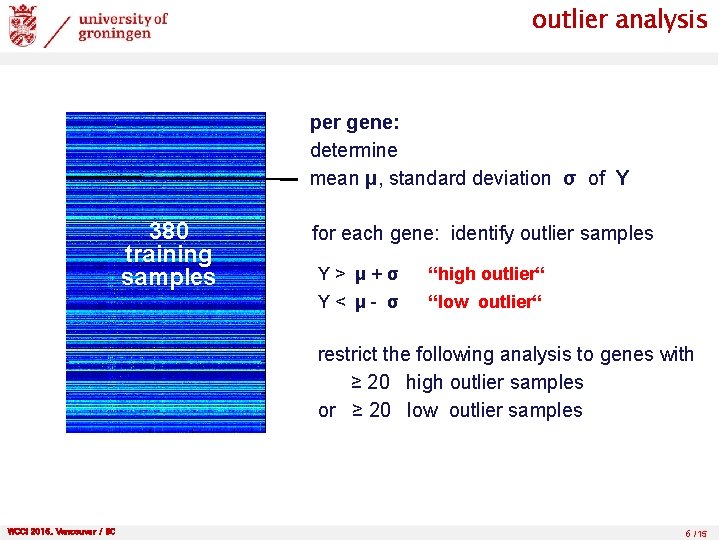
outlier analysis per gene: determine mean μ, standard deviation σ of Y 380 training samples for each gene: identify outlier samples Y> μ+σ “high outlier“ Y< μ- σ “low outlier“ restrict the following analysis to genes with ≥ 20 high outlier samples or ≥ 20 low outlier samples WCCI 2016, Vancouver / BC 6 /15

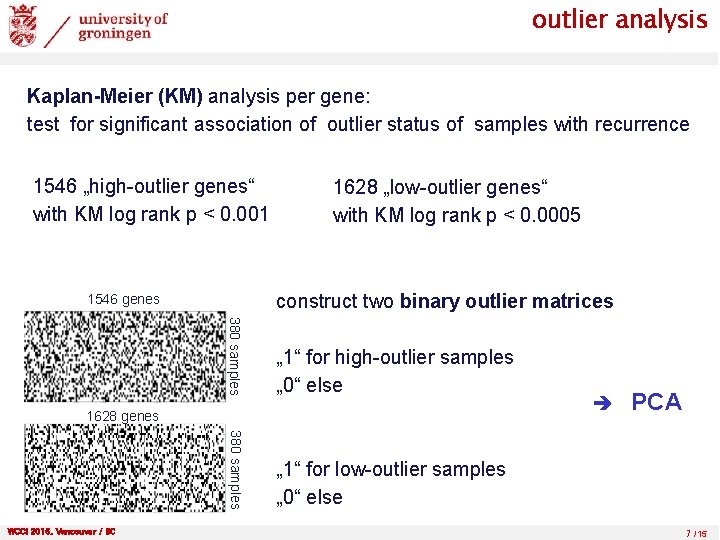
outlier analysis Kaplan-Meier (KM) analysis per gene: test for significant association of outlier status of samples with recurrence 1546 „high-outlier genes“ with KM log rank p < 0. 001 1628 „low-outlier genes“ with KM log rank p < 0. 0005 construct two binary outlier matrices 1546 genes 380 samples „ 1“ for high-outlier samples „ 0“ else 380 samples „ 1“ for low-outlier samples „ 0“ else 1628 genes WCCI 2016, Vancouver / BC PCA 7 /15

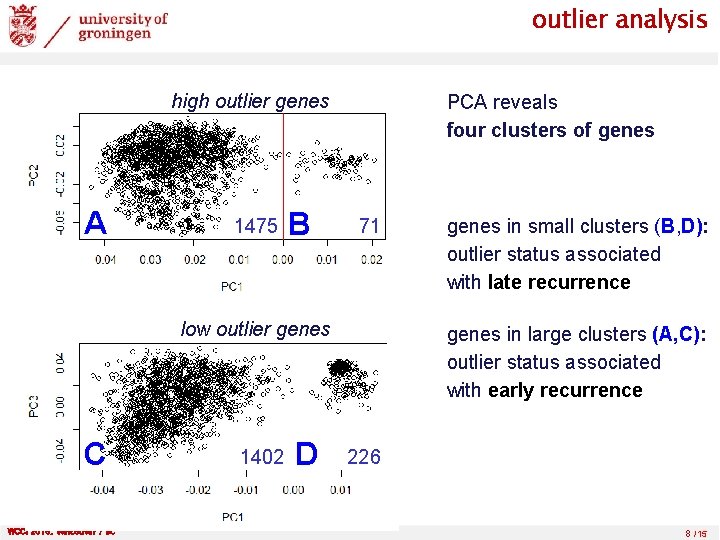
outlier analysis high outlier genes A 1475 B PCA reveals four clusters of genes 71 low outlier genes C WCCI 2016, Vancouver / BC 1402 D genes in small clusters (B, D): outlier status associated with late recurrence genes in large clusters (A, C): outlier status associated with early recurrence 226 8 /15

recurrence risk score top 20 genes (by KM p-value) from each cluster A, B, C, D reference set of 80 genes for each sample: - determine outlier status with respect to the 80 genes (Y >? < μ ± σ ) - add up contributions per gene - 1 if the sample is outlier w. r. t. to a gene in A or C (early rec. ) 0 if the sample is not an outlier w. r. t. the gene + 1 if the sample is outlier w. r. t. to a gene in B or D (late rec. ) recurrence risk score - 40 ≤ R ≤ + 40 observe: median = 2 over the 380 training samples crisp classification w. r. t. recurrence risk: high risk (early recurrence) if R < 2 low risk (late recurrence) if R ≥ 2 WCCI 2016, Vancouver / BC 9 /15

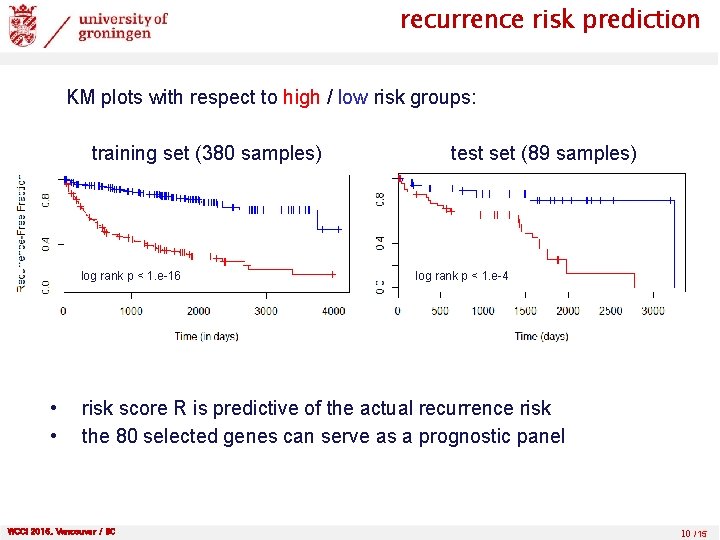
recurrence risk prediction KM plots with respect to high / low risk groups: training set (380 samples) log rank p < 1. e-16 • • test set (89 samples) log rank p < 1. e-4 risk score R is predictive of the actual recurrence risk the 80 selected genes can serve as a prognostic panel WCCI 2016, Vancouver / BC 10 /15

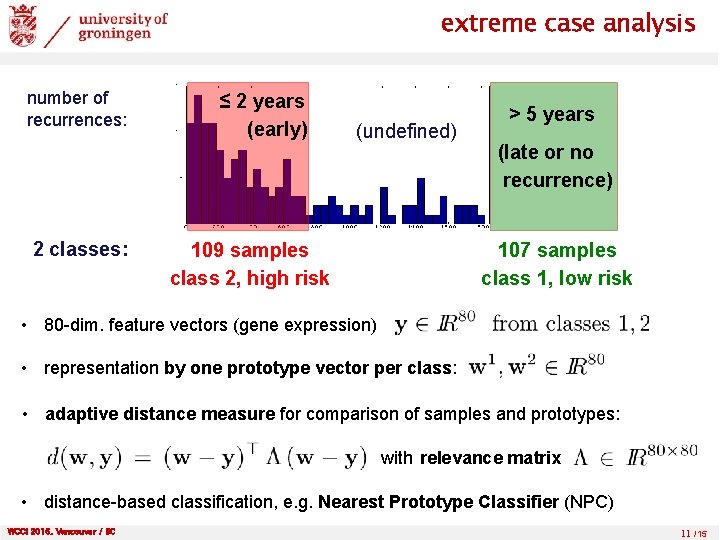
extreme case analysis number of recurrences: 2 classes: ≤ 2 years (early) (undefined) 109 samples class 2, high risk > 5 years (late or no recurrence) 107 samples class 1, low risk • 80 -dim. feature vectors (gene expression) • representation by one prototype vector per class: • adaptive distance measure for comparison of samples and prototypes: with relevance matrix • distance-based classification, e. g. Nearest Prototype Classifier (NPC) WCCI 2016, Vancouver / BC 11 /15

GMLVQ classifier low expression | high expression Generalized Matrix Relevance Learning Vector Quantization (GMLVQ) training of prototypes and relevance matrix = minimization of an appropriate cost function with respect to performance on labeled training set diagonal elements of Λ components of A WCCI 2016, Vancouver / BC B C D A B C D 12 /15

GMLVQ classifier ROC of GMLVQ classifier (Leave-One-Out of the 216 extreme samples) log rank p < 1. e-7 KM plot w. r. t. all 469 samples ( L-1 -O for 216 samples, plus 253 undefined ) WCCI 2016, Vancouver / BC 13 /15

extreme case analysis (107+109 samples) GMLVQ classifier Risk score classifier - AUC=0. 84 R=2 WCCI 2016, Vancouver / BC 14 /15

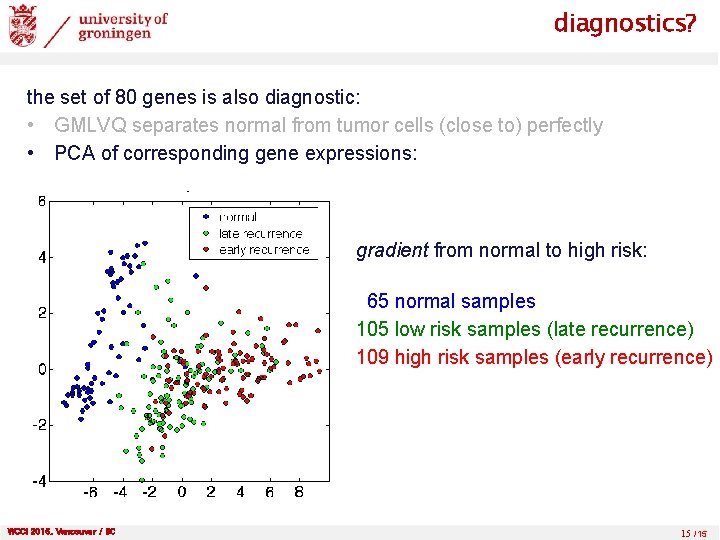
diagnostics? the set of 80 genes is also diagnostic: • GMLVQ separates normal from tumor cells (close to) perfectly • PCA of corresponding gene expressions: gradient from normal to high risk: 65 normal samples 105 low risk samples (late recurrence) 109 high risk samples (early recurrence) WCCI 2016, Vancouver / BC 15 /15

remarks and open questions • prospective studies required with respect to use as an assay • 80 genes do not necessarily reflect biological mechanisms compare, e. g. , with known pathways / modules of genes • GMLVQ suggests an even smaller panel of prognostic genes (12? ) identify a minimum panel for diagnostics and prognostics • can the performance be improved further ? study more sophisticated classifier systems include further clinical information (diet, life style, family history, … ) • more direct, multivariate identification of relevant genes ? e. g. PCA+GMLVQ and back-transform easy-to-use GMLVQ-classifier: www. cs. rug. nl/~biehl/gmlvq WCCI 2016, Vancouver / BC 16 /15