Pairwise sequence alignment practice 1 Manual Work Write






- Slides: 16

Pairwise sequence alignment (practice) 1

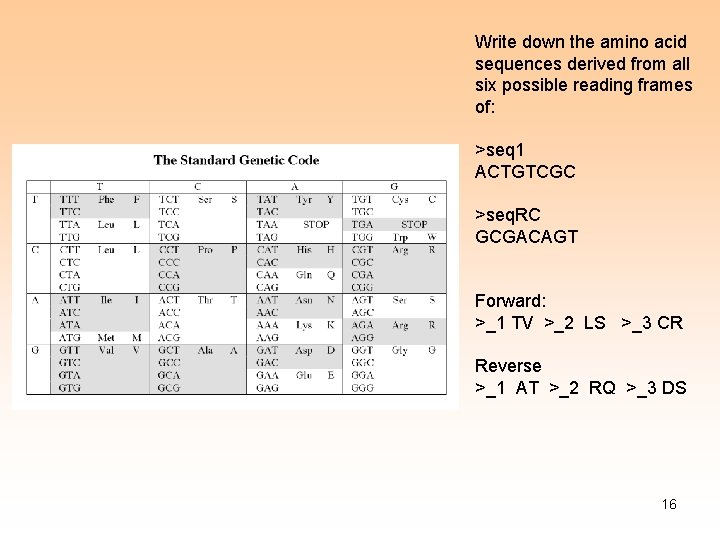
Manual Work: Write down the amino acid sequences derived from all six possible reading frames of: >seq 1 ACTGTCGC 2

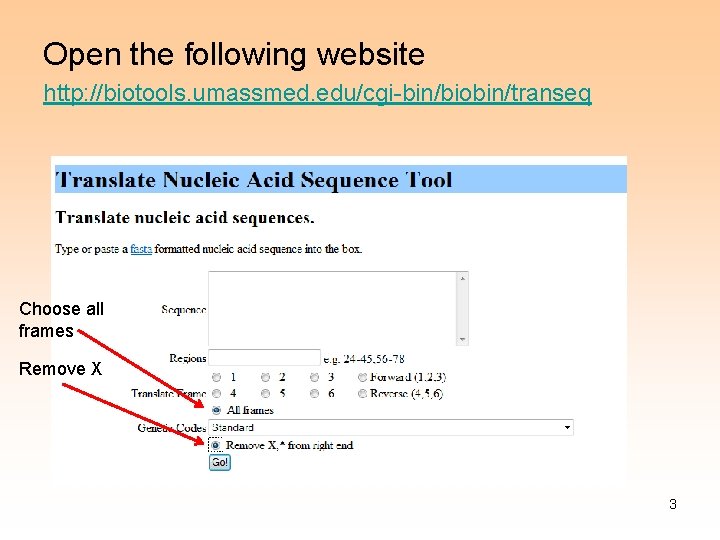
Open the following website http: //biotools. umassmed. edu/cgi-bin/biobin/transeq Choose all frames Remove X 3

Translate all possible reading frames of the following sequence TATGTCTCTCACCAACAAGAACGTCGTTTTCGTGGCCGGTCTGGGCGGCATTGGCCTGGAC ACCAGCCGGGAGTTGGTCAAGCGTAATCTGAAGAACCTGGTCATCCTGGATCGCATTGAC AATCCGGCTGCCATTGCCGAACTGAAGGCAATCCCAAGGTGACCATCACCTTCTAT CCCTACGATGTGACTGTGCCCGTCGCTGAGACCACCAAGCTCCTGAAGACCATCTTTGCC CAGGTGAAGACAATCGATGTCCTGATCAACGGTGCTGGCATCCTGGACGATCATCAGATC GAGCGCACCATTGCCGTTAACTACACGGGCCTGGTCAACACCACCACAGCCATTCTGGAC TTCTGGGACAAGCGCAAGGGCGGCCCAGGCGGCATCATTTGCAACATTGGCTCCGTCACC GGTTTCAATGCCATCTACCAGGTGCCCGTTTACTCCGGCTCCAAGGCGGCGGTGGTTAAC TTTACCTCCTCCCTGGCGAAACTGGCTCCCATTACTGGTGTCACTGCTTACACTGTCAAT CCTGGCATCACCAGGACCACTCTGGTCCACAAATTCAACTCGTGGCTGGATGTGGAGCCC CGTGTGGCGGAGAAGCTGCTCGAGCATCCCAGACCTCTCAGCAGTGCGCCGAGAAC TTTGTGAAGGCCATTGAGCTGAACAAGAACGGTGCCATCTGGAAGTTGGATTTGGGCACC TTGGAGCCCATCACATGGACCCAGCACTGGGACTCGGGCATCTAA Which reading frame(s) is(are) likely to be the true reading frame(s)? 4


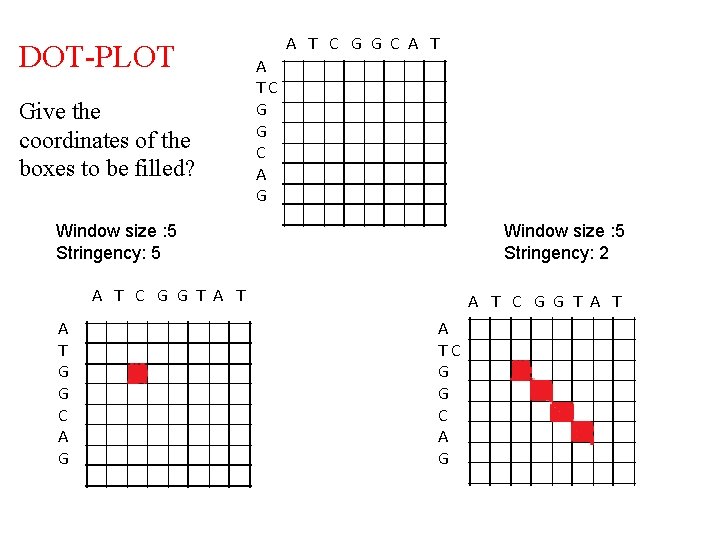
DOT-PLOT Give the coordinates of the boxes to be filled? A T C G G C A TC G G C A G Window size : 5 Stringency: 5 Window size : 5 Stringency: 2 A T C G G T A T A TC G G C A G

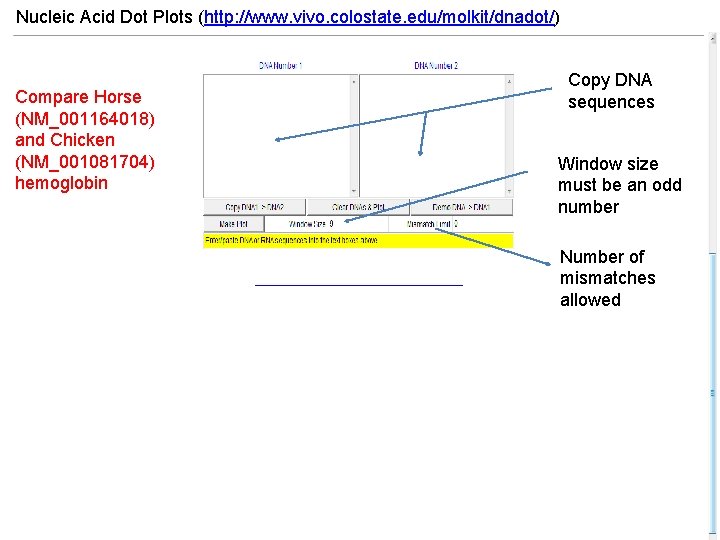
Nucleic Acid Dot Plots (http: //www. vivo. colostate. edu/molkit/dnadot/) Compare Horse (NM_001164018) and Chicken (NM_001081704) hemoglobin Copy DNA sequences Window size must be an odd number Number of mismatches allowed


Use the Dotplot to compare chicken ovomucoid (NM_001112662) to itself GCACCGGCAGCCGCCTGCAGAGCCGGGCAGTACCTCACCATGGCAGGCGTCTTCGTGCTGTTCT CTTTCGTGCTTTGTGGCTTCCTCCCAGATGCTGCCTTTGGGGCTGAGGTGGACTGCAGTAGGTTTCCCAA CGCTACAGACAAGGCAAAGATGTATTGGTTTGCAACAAGGACCTCCGCCCCATCTGTGGTACCGAT GGAGTCACTTACACCAACGATTGCTGTGTGCCTACAGCATAGAATTTGGAACCAATATCAGCAAAG AGCACGATGGAGAATGCAAGGAAACTGTTCCTATGAACTGCAGTAGTTATGCCAACACGACAAGCGAGGA CGGAAAAGTGATGGTCCTCTGCAACAGGGCCTTCAACCCCGTCTGTGGTACTGATGGAGTCACCTACGAC AATGAGTGTCTGCTGTGTGCCCACAAAGTAGAGCAGGGGGCCAGCGTTGACAAGAGGCATGATGGTGGAT GTAGGAACTTGCTGCTGTTGACTGCAGCGAGTACCCTAAGCCTGACTGCACGGCAGAAGACC TCTCTGTGGCTCCGACAACAAAACATATGGCAACAAGTGCAACTTCTGCAATGCAGTCGTGGAAAGCAAC GGGACTCTCACTTTAAGCCATTTTGGAAAATGCTGAATATCAGAGCTGAGAGAATTCACCACAGGATCCC CACTGGCGAATCCCAGCGAGAGGTCTCACCTCGGTTCATCTCGCACTCTGGGGAGCTCACTCCCG ATTTTCTCAATAAACTAAATCAGCAACAAAAA

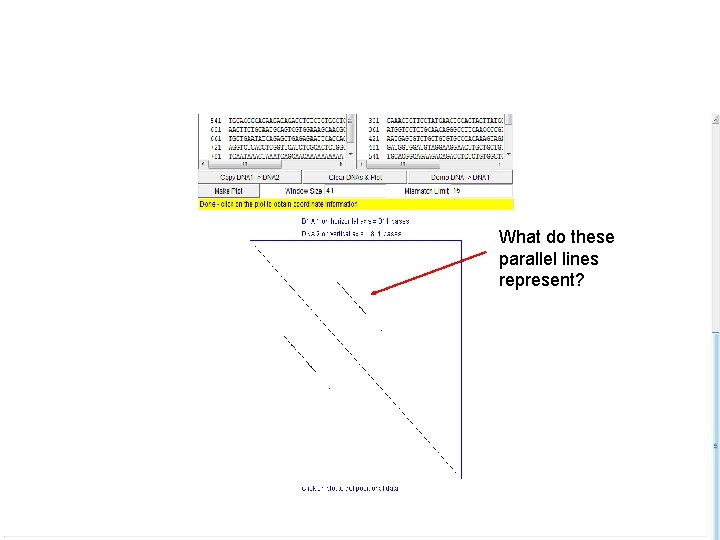
What do these parallel lines represent?

LALIGN - finds multiple matching subsegments in two sequences Part of the FASTA package of sequence analysis program. Lalign - compares two protein or DNA sequences for local or global similarity and shows the local sequence alignments. http: //www. ch. embnet. org/software/LALIGN_form. html 11

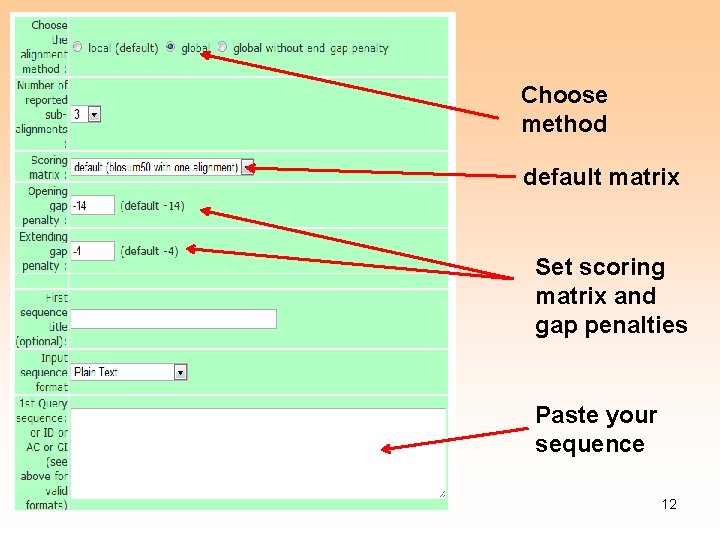
Choose method default matrix Set scoring matrix and gap penalties Paste your sequence 12

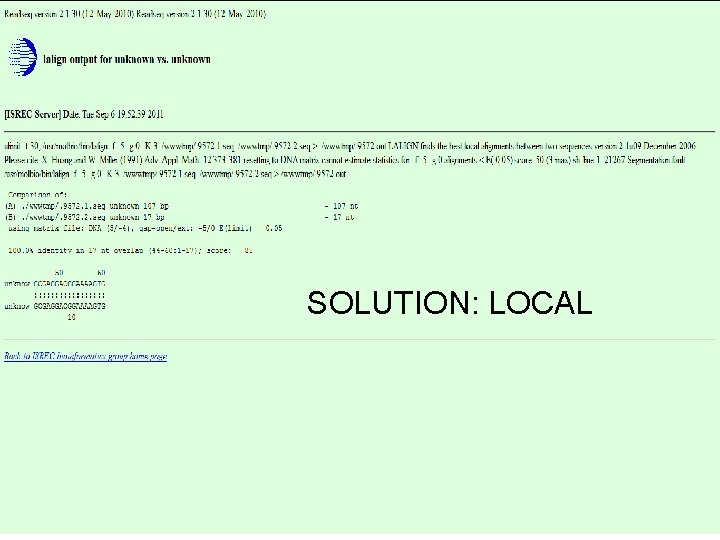
Open http: //www. ch. embnet. org/software/LALIGN_form. html Use the sequences below and perform a global alignment with 5 “Opening gap penalty” and 0 “Extending gap penalty”. >seq 1 GCGACTGTTCCTATGAACTGCAGTAGTTATGCCAACACG ACAAGCGAGGACGGAAAAGTGAGTCTGTGGTACTGATGG AGTCACCTACGACGCGAGGACGCCAGGTG >seq 2 GCGAGGACGGAAAAGTG 13

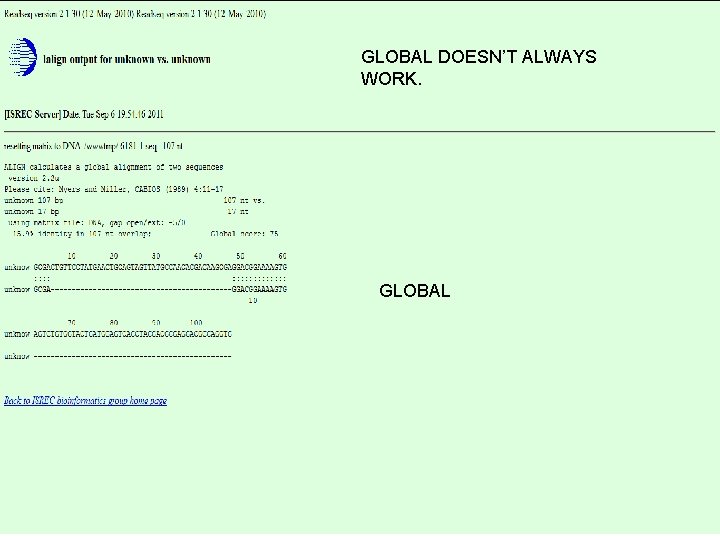
GLOBAL DOESN’T ALWAYS WORK. GLOBAL 14

SOLUTION: LOCAL 15

Write down the amino acid sequences derived from all six possible reading frames of: >seq 1 ACTGTCGC >seq. RC GCGACAGT Forward: >_1 TV >_2 LS >_3 CR Reverse >_1 AT >_2 RQ >_3 DS 16