New SNPs Sift Polyphen etc SIFT predicting amino

![Gene-Gene Interactions "Model. . . erythrocytes of [glutathione peroxidase] GPX 1*2 heterozygotes should be Gene-Gene Interactions "Model. . . erythrocytes of [glutathione peroxidase] GPX 1*2 heterozygotes should be](https://slidetodoc.com/presentation_image_h2/acf1222f00d963ce467c202e2caf63ee/image-4.jpg)

- Slides: 20

New SNPs: Sift, Polyphen, etc. SIFT: predicting amino acid changes that affect protein function Pauline C. Ng and Steven Henikoff, Nucleic Acids Research, 2003, Vol. 3 1, No. 13 3812 -3814

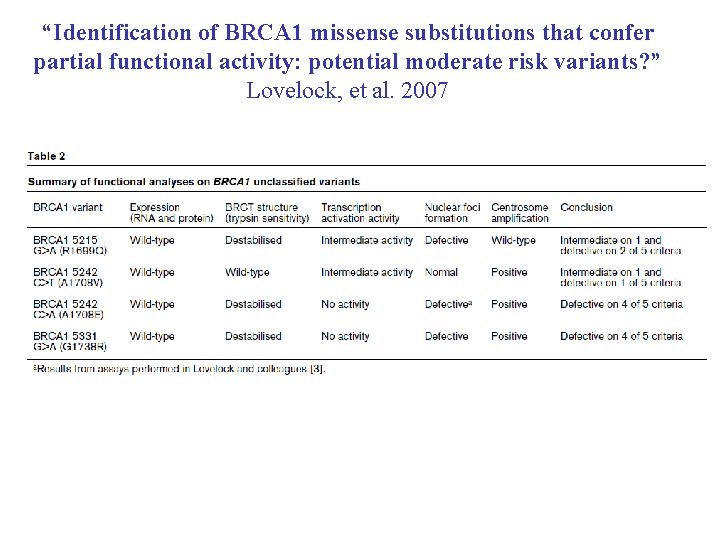
“Identification of BRCA 1 missense substitutions that confer partial functional activity: potential moderate risk variants? ” Lovelock, et al. 2007

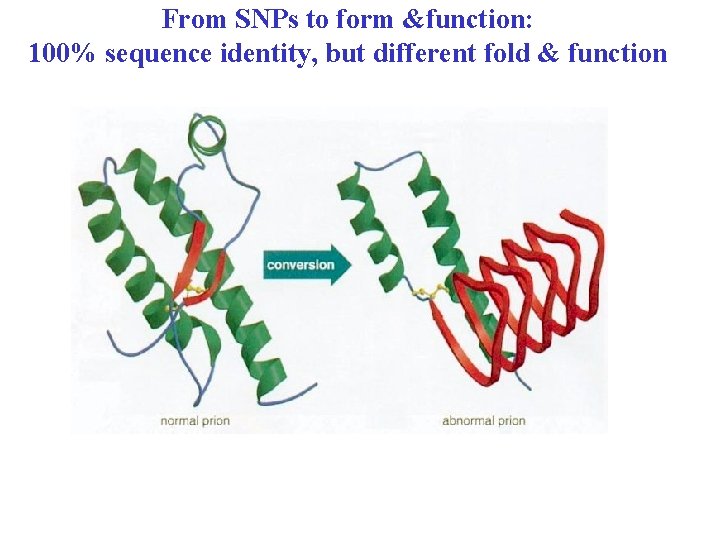
From SNPs to form &function: 100% sequence identity, but different fold & function
![GeneGene Interactions Model erythrocytes of glutathione peroxidase GPX 12 heterozygotes should be Gene-Gene Interactions "Model. . . erythrocytes of [glutathione peroxidase] GPX 1*2 heterozygotes should be](https://slidetodoc.com/presentation_image_h2/acf1222f00d963ce467c202e2caf63ee/image-4.jpg)
Gene-Gene Interactions "Model. . . erythrocytes of [glutathione peroxidase] GPX 1*2 heterozygotes should be more efficient in sheltering the cell membrane from irreversible oxidation and binding of hemoglobin caused by the oxidant stress exerted by Plasmodium falciparum. . . we observed a clear trend toward a dissociation between the HBB*A/*S and GPX 1*2/*1 genotypes in the overall data. " Destro-Bisol et al. Hum Biol 1999; 71: 315 -32. (Pub) 4

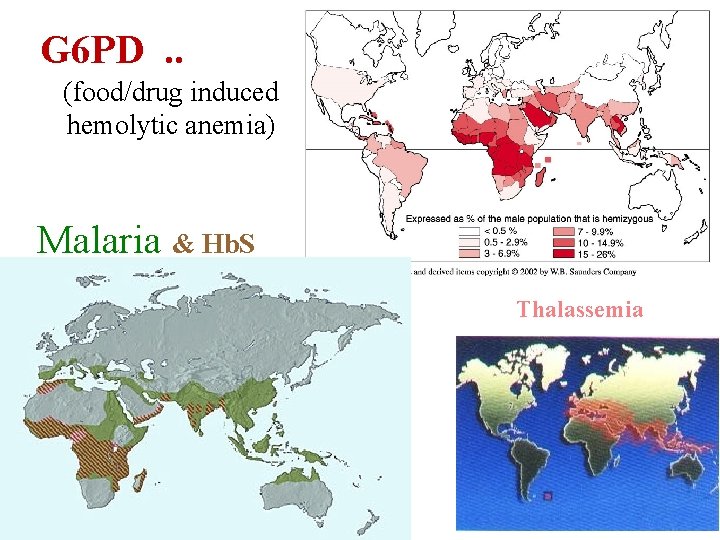
G 6 PD. . (food/drug induced hemolytic anemia) Malaria & Hb. S Thalassemia

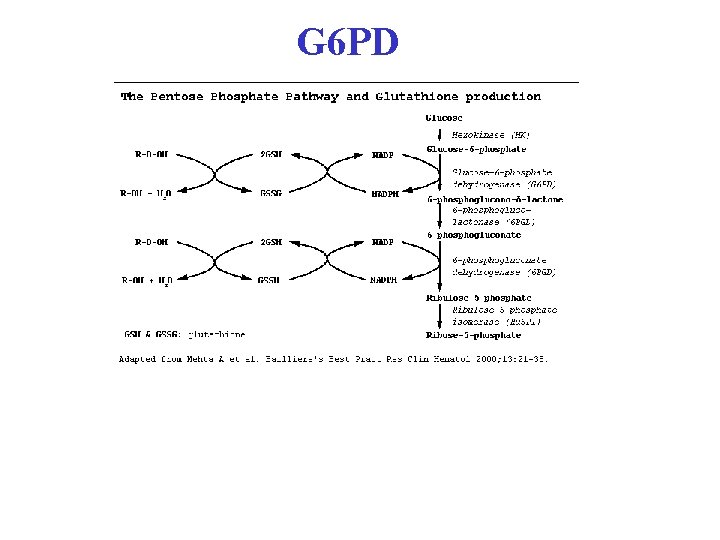
G 6 PD

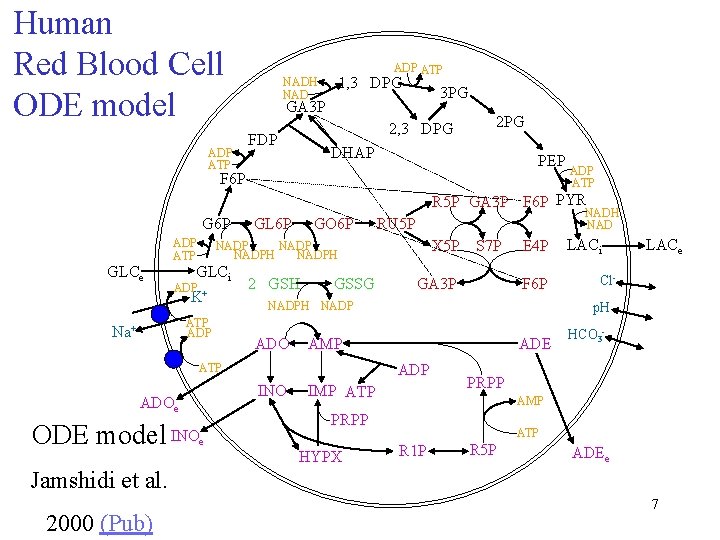
Human Red Blood Cell ODE model ADP ATP NADH NAD ADP ATP 1, 3 DPG 3 PG GA 3 P 2, 3 DPG FDP 2 PG DHAP PEP F 6 P ADP ATP R 5 P GA 3 P F 6 P PYR G 6 P GLCe ADP ATP GO 6 P GLCi ATP ADP 2 GSH GSSG X 5 P S 7 P ADO F 6 P AMP ADE IMP ATP HYPX LACe Cl- HCO 3 - PRPP AMP PRPP e LACi p. H ADP INO ADOe GA 3 P E 4 P NADPH NADP ATP ODE model INO NADH NAD RU 5 P NADPH ADP + K Na+ GL 6 P ATP R 1 P R 5 P ADEe Jamshidi et al. 2000 (Pub) 7

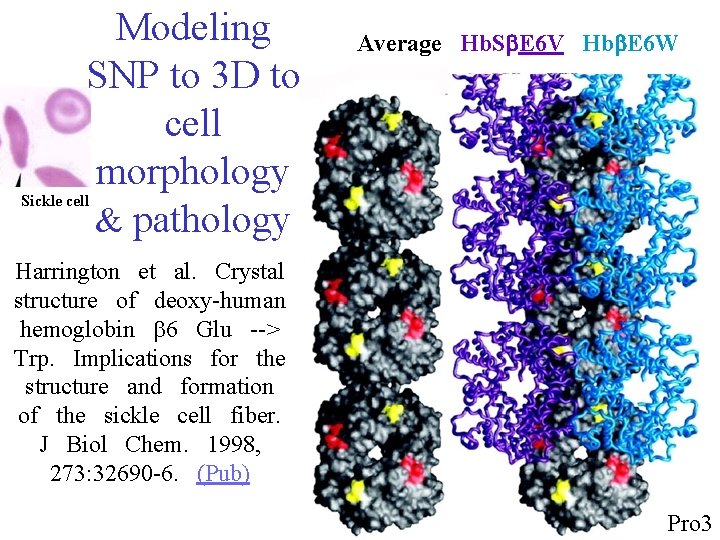
Modeling SNP to 3 D to cell morphology & pathology Average Hb. Sb. E 6 V Hbb. E 6 W Sickle cell Harrington et al. Crystal structure of deoxy-human hemoglobin b 6 Glu --> Trp. Implications for the structure and formation of the sickle cell fiber. J Biol Chem. 1998, 273: 32690 -6. (Pub) 8 Pro 3

Red Blood Cell Function • Transport O 2 from lungs to tissues – using hemoglobin to carry the O 2 • Hemoglobin is maintained in its functional state (reduced) by the metabolic machinery • Cell membrane separates the internal environment from the external environment – subject to physicochemical constraints • Electroneutrality, Osmotic balance – Cause of the imbalance: • impermeable polyions inside the cell – hemoglobin, organic phosphates 9

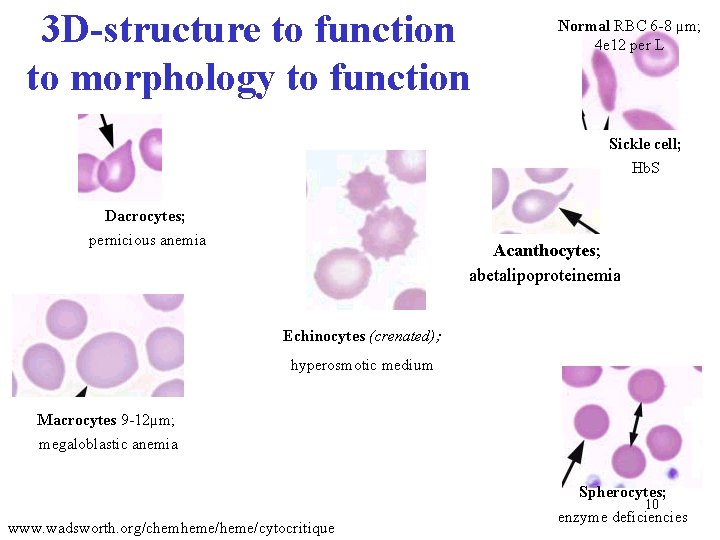
3 D-structure to function to morphology to function Normal RBC 6 -8 µm; 4 e 12 per L Sickle cell; Hb. S Dacrocytes; pernicious anemia Acanthocytes; abetalipoproteinemia Echinocytes (crenated); hyperosmotic medium Macrocytes 9 -12µm; megaloblastic anemia Spherocytes; 10 www. wadsworth. org/chemheme/cytocritique enzyme deficiencies

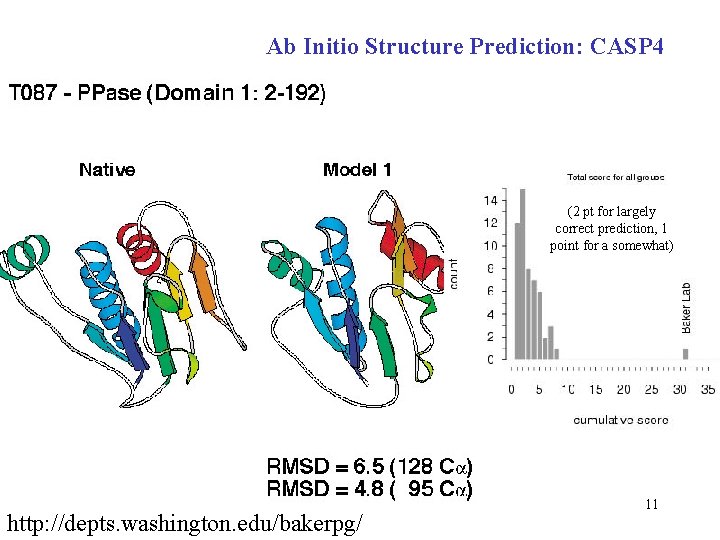
Ab Initio Structure Prediction: CASP 4 (2 pt for largely correct prediction, 1 point for a somewhat) http: //depts. washington. edu/bakerpg/ 11

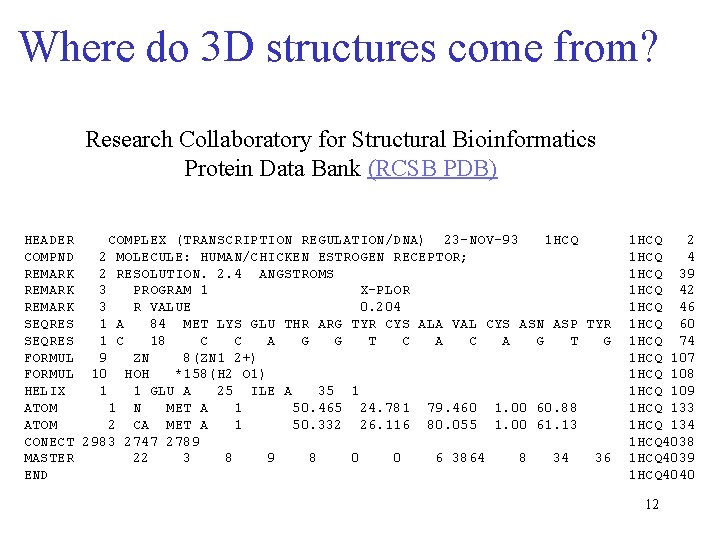
Where do 3 D structures come from? Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) HEADER COMPLEX (TRANSCRIPTION REGULATION/DNA) 23 -NOV-93 1 HCQ COMPND 2 MOLECULE: HUMAN/CHICKEN ESTROGEN RECEPTOR; REMARK 2 RESOLUTION. 2. 4 ANGSTROMS REMARK 3 PROGRAM 1 X-PLOR REMARK 3 R VALUE 0. 204 SEQRES 1 A 84 MET LYS GLU THR ARG TYR CYS ALA VAL CYS ASN ASP TYR SEQRES 1 C 18 C C A G G T C A G T G FORMUL 9 ZN 8(ZN 1 2+) FORMUL 10 HOH *158(H 2 O 1) HELIX 1 1 GLU A 25 ILE A 35 1 ATOM 1 N MET A 1 50. 465 24. 781 79. 460 1. 00 60. 88 ATOM 2 CA MET A 1 50. 332 26. 116 80. 055 1. 00 61. 13 CONECT 2983 2747 2789 MASTER 22 3 8 9 8 0 0 6 3864 8 34 36 END 1 HCQ 2 1 HCQ 4 1 HCQ 39 1 HCQ 42 1 HCQ 46 1 HCQ 60 1 HCQ 74 1 HCQ 107 1 HCQ 108 1 HCQ 109 1 HCQ 133 1 HCQ 134 1 HCQ 4038 1 HCQ 4039 1 HCQ 4040 12

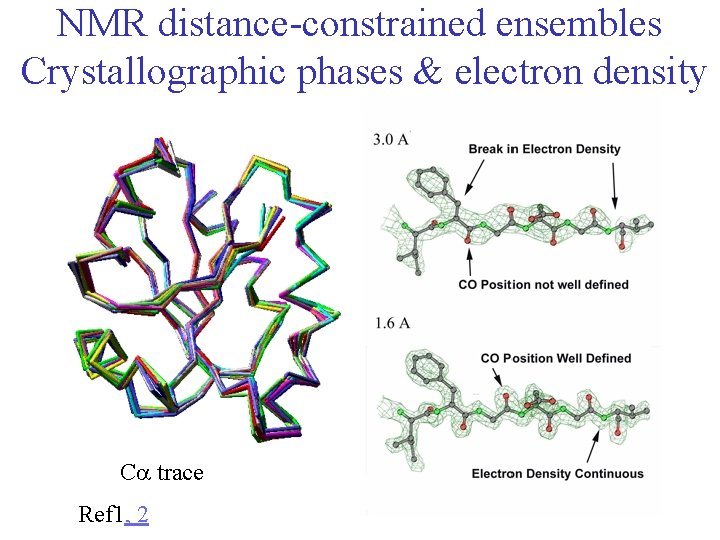
NMR distance-constrained ensembles Crystallographic phases & electron density Ca trace Ref 1, 2 13

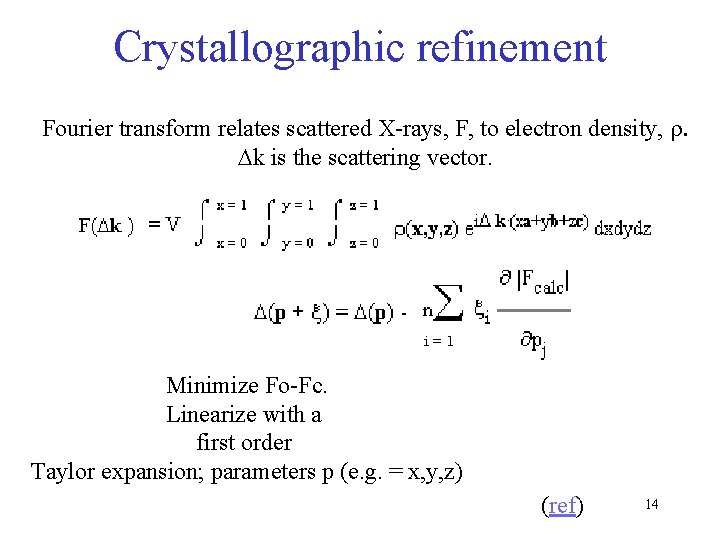
Crystallographic refinement Fourier transform relates scattered X-rays, F, to electron density, r. Dk is the scattering vector. Minimize Fo-Fc. Linearize with a first order Taylor expansion; parameters p (e. g. = x, y, z) (ref) 14

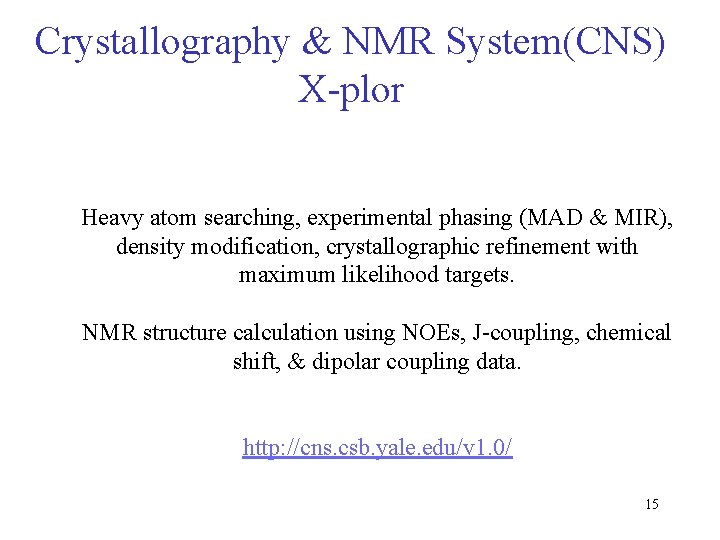
Crystallography & NMR System(CNS) X-plor Heavy atom searching, experimental phasing (MAD & MIR), density modification, crystallographic refinement with maximum likelihood targets. NMR structure calculation using NOEs, J-coupling, chemical shift, & dipolar coupling data. http: //cns. csb. yale. edu/v 1. 0/ 15

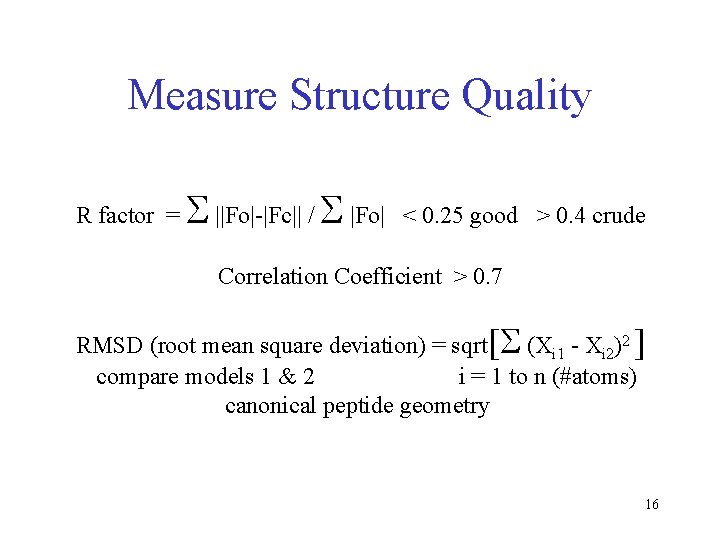
Measure Structure Quality R factor = S ||Fo|-|Fc|| / S |Fo| < 0. 25 good > 0. 4 crude Correlation Coefficient > 0. 7 RMSD (root mean square deviation) = sqrt[S (Xi 1 - Xi 2)2 ] compare models 1 & 2 i = 1 to n (#atoms) canonical peptide geometry 16

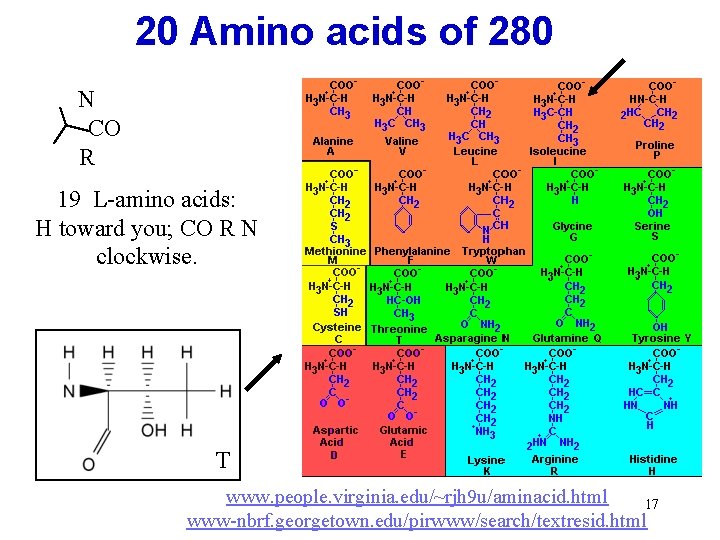
20 Amino acids of 280 N CO R 19 L-amino acids: H toward you; CO R N clockwise. T www. people. virginia. edu/~rjh 9 u/aminacid. html 17 www-nbrf. georgetown. edu/pirwww/search/textresid. html

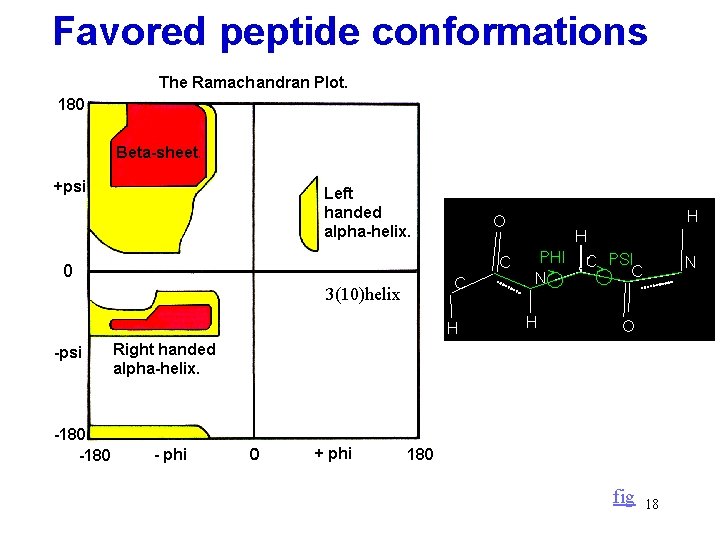
Favored peptide conformations 3(10)helix fig 18

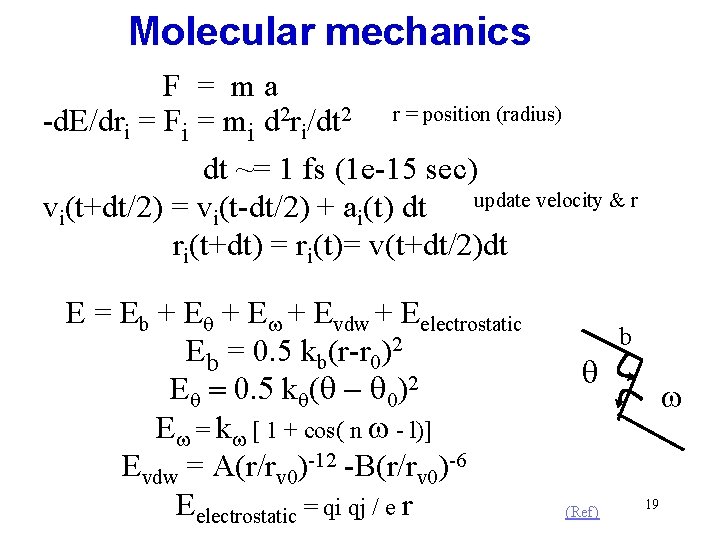
Molecular mechanics F = ma -d. E/dri = Fi = mi d 2 ri/dt 2 r = position (radius) dt ~= 1 fs (1 e-15 sec) update velocity & r vi(t+dt/2) = vi(t-dt/2) + ai(t) dt ri(t+dt) = ri(t)= v(t+dt/2)dt E = Eb + Eq + Ew + Evdw + Eelectrostatic Eb = 0. 5 kb(r-r 0)2 Eq = 0. 5 kq(q - q 0)2 Ew = kw [ 1 + cos( n w - l)] Evdw = A(r/rv 0)-12 -B(r/rv 0)-6 Eelectrostatic = qi qj / e r b q (Ref) w 19
