MESA Family Genetics Committee Report Jerome I Rotter


































- Slides: 52

MESA Family Genetics Committee Report Jerome I. Rotter Monday, Sept. 18, 2006 1

Outline • MESA and MESA Family – – – “Prior” Candidate Gene Analyses Phase 2 genotyping Phase 1 genotyping and analysis Gene. Pages (Mychaleckyj presentation) Publications Committee (Taylor presentation) • MESA Family – Recruitment status (Operations Committee) – Planned analyses (Raffel and Pankow presentations) – Genome-wide linkage analysis • Operational issues: MESA Genetics Committee • CARE • NIH RFI on Data Sharing 2

Overview of Progress MESA and MESA Family • 6 manuscripts from “prior” candidate genes analyses in or pending publication (prior to MESA Family large-scale genotyping effort) • Phase 2 genotyping project underway • Analysis of Phase 1 candidate genes • Genepage development underway • Proposed genetic specific P&P committee for candidate gene proposals (parallel to MESA P&P) 3

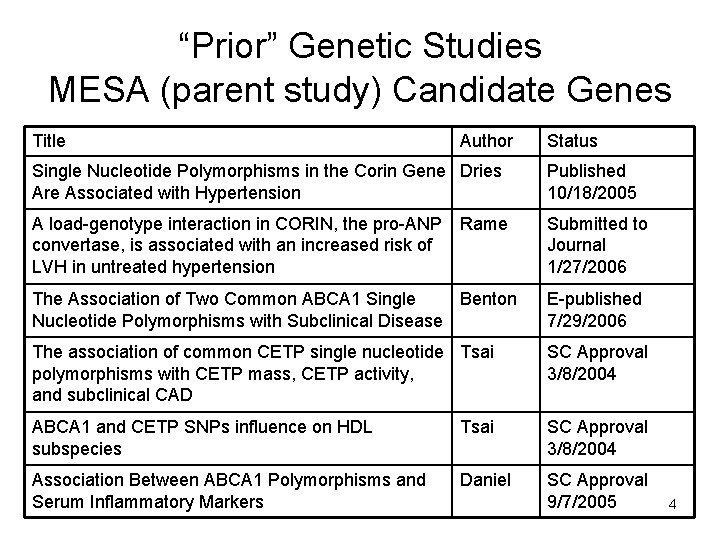
“Prior” Genetic Studies MESA (parent study) Candidate Genes Title Author Status Single Nucleotide Polymorphisms in the Corin Gene Dries Are Associated with Hypertension Published 10/18/2005 A load-genotype interaction in CORIN, the pro-ANP Rame convertase, is associated with an increased risk of LVH in untreated hypertension Submitted to Journal 1/27/2006 The Association of Two Common ABCA 1 Single Benton Nucleotide Polymorphisms with Subclinical Disease E-published 7/29/2006 The association of common CETP single nucleotide Tsai polymorphisms with CETP mass, CETP activity, and subclinical CAD SC Approval 3/8/2004 ABCA 1 and CETP SNPs influence on HDL subspecies Tsai SC Approval 3/8/2004 Association Between ABCA 1 Polymorphisms and Serum Inflammatory Markers Daniel SC Approval 9/7/2005 4

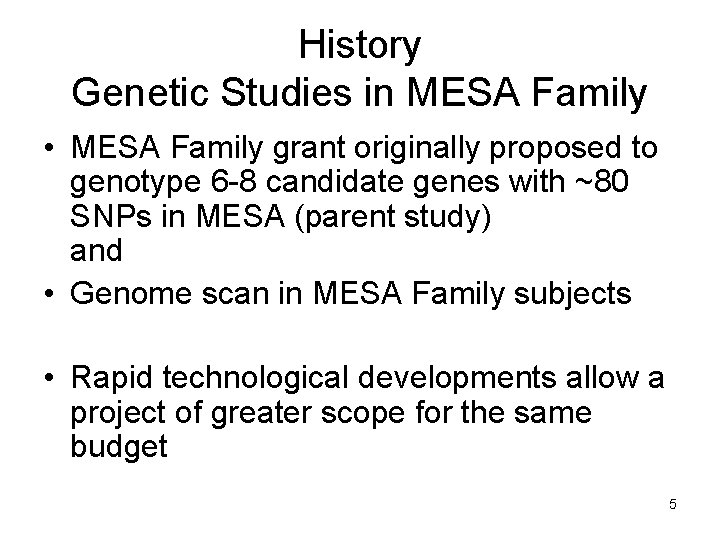
History Genetic Studies in MESA Family • MESA Family grant originally proposed to genotype 6 -8 candidate genes with ~80 SNPs in MESA (parent study) and • Genome scan in MESA Family subjects • Rapid technological developments allow a project of greater scope for the same budget 5

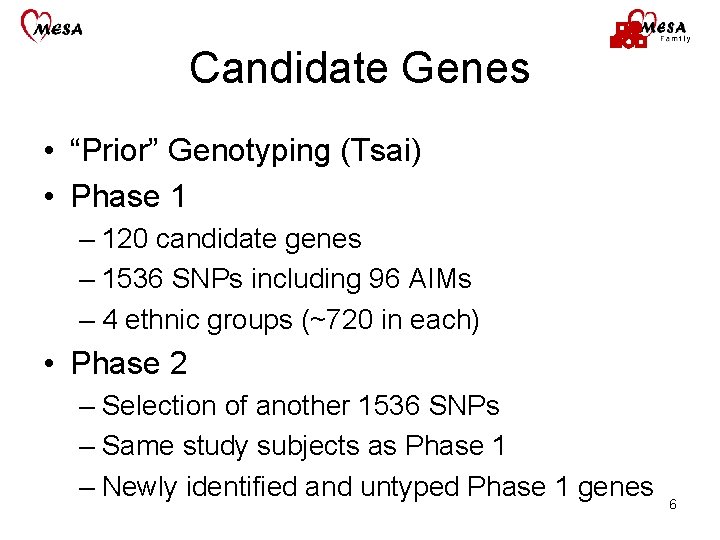
Candidate Genes • “Prior” Genotyping (Tsai) • Phase 1 – 120 candidate genes – 1536 SNPs including 96 AIMs – 4 ethnic groups (~720 in each) • Phase 2 – Selection of another 1536 SNPs – Same study subjects as Phase 1 – Newly identified and untyped Phase 1 genes 6

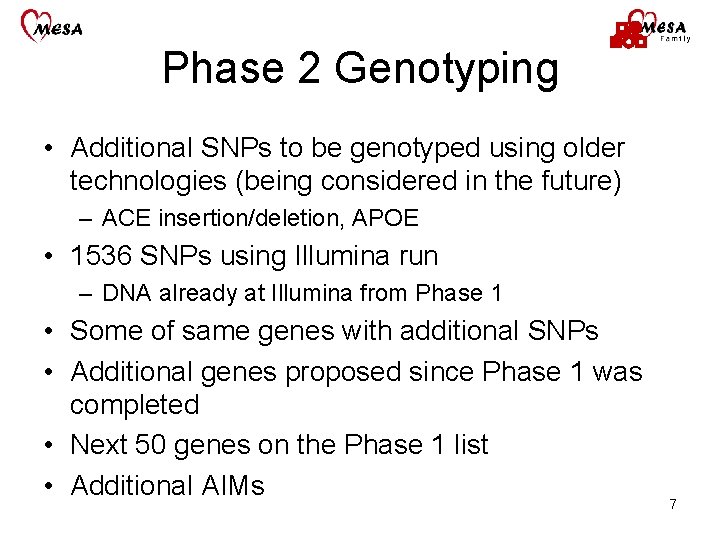
Phase 2 Genotyping • Additional SNPs to be genotyped using older technologies (being considered in the future) – ACE insertion/deletion, APOE • 1536 SNPs using Illumina run – DNA already at Illumina from Phase 1 • Some of same genes with additional SNPs • Additional genes proposed since Phase 1 was completed • Next 50 genes on the Phase 1 list • Additional AIMs 7

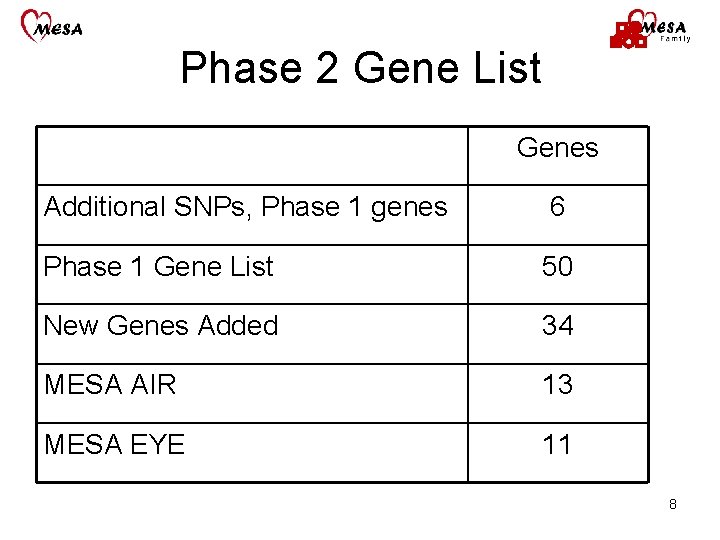
Phase 2 Gene List Genes Additional SNPs, Phase 1 genes 6 Phase 1 Gene List 50 New Genes Added 34 MESA AIR 13 MESA EYE 11 8

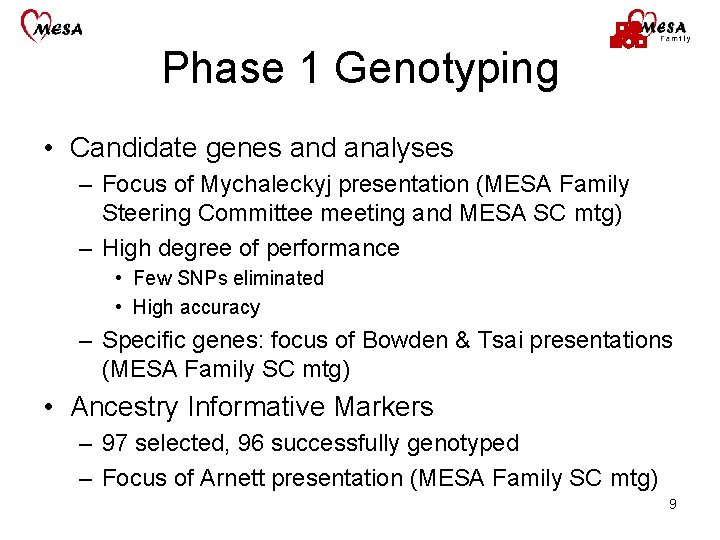
Phase 1 Genotyping • Candidate genes and analyses – Focus of Mychaleckyj presentation (MESA Family Steering Committee meeting and MESA SC mtg) – High degree of performance • Few SNPs eliminated • High accuracy – Specific genes: focus of Bowden & Tsai presentations (MESA Family SC mtg) • Ancestry Informative Markers – 97 selected, 96 successfully genotyped – Focus of Arnett presentation (MESA Family SC mtg) 9

Genome Wide Linkage Scan • Originally planned to utilize Mammalian Genotyping Service • Issue: no more Mammalian Genotyping Service • Plan: Supplemental funding request 10

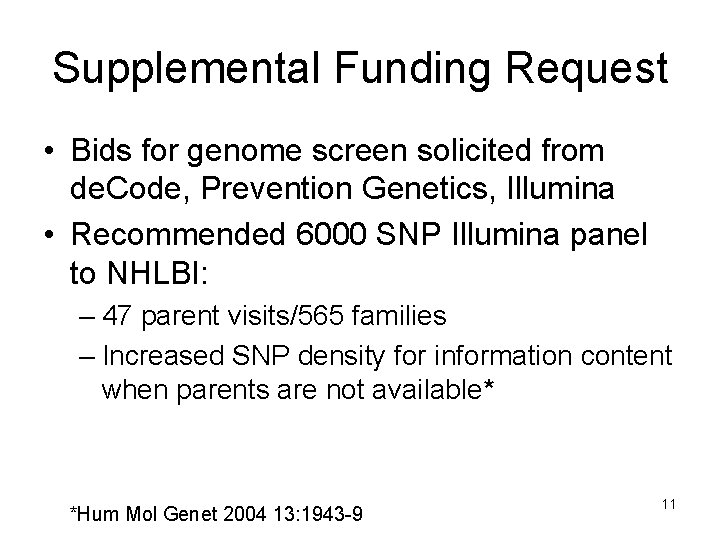
Supplemental Funding Request • Bids for genome screen solicited from de. Code, Prevention Genetics, Illumina • Recommended 6000 SNP Illumina panel to NHLBI: – 47 parent visits/565 families – Increased SNP density for information content when parents are not available* *Hum Mol Genet 2004 13: 1943 -9 11

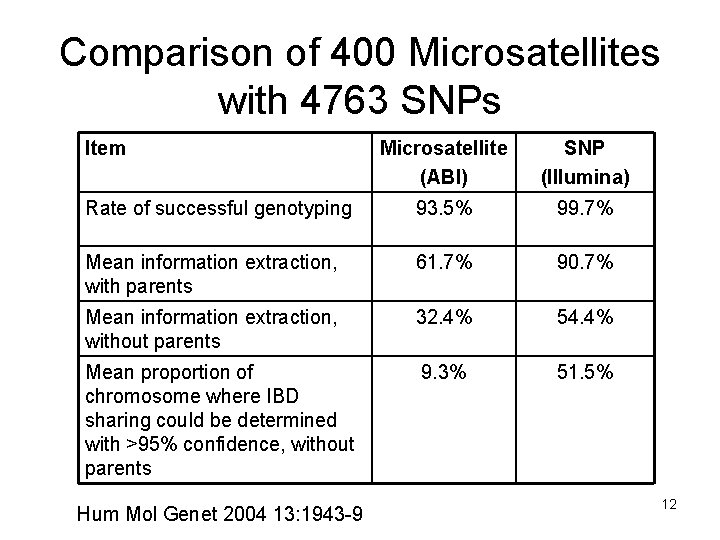
Comparison of 400 Microsatellites with 4763 SNPs Item Microsatellite (ABI) SNP (Illumina) Rate of successful genotyping 93. 5% 99. 7% Mean information extraction, with parents 61. 7% 90. 7% Mean information extraction, without parents 32. 4% 54. 4% Mean proportion of chromosome where IBD sharing could be determined with >95% confidence, without parents 9. 3% 51. 5% Hum Mol Genet 2004 13: 1943 -9 12

MESA Genetics Operational Issues • Handling small genotyping projects (Tsai, Taylor, Bowden subcommittee proposal) • Handling collaborations when genes/ markers are unknown at onset 13

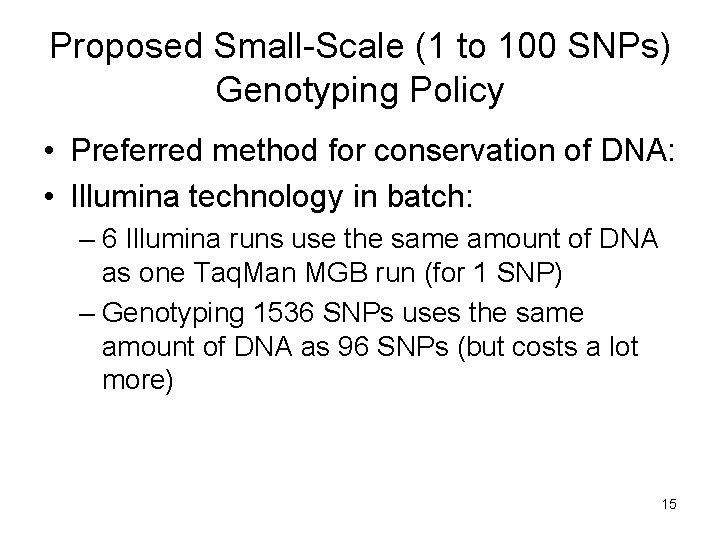
Proposed Small-Scale (1 to 100 SNPs) Genotyping Policy • Sending DNA of MESA out to investigators is an inefficient use of the available resource for genotyping of only a few (1 to 100) SNPs: – Amount of DNA is finite and large amounts will be wasted – Too labor-intensive for a large number of requests • Therefore genotyping will be performed at one of the three MESA genotyping laboratories by Bowden, Tsai, or Taylor • Investigator(s) will be responsible for costs 14

Proposed Small-Scale (1 to 100 SNPs) Genotyping Policy • Preferred method for conservation of DNA: • Illumina technology in batch: – 6 Illumina runs use the same amount of DNA as one Taq. Man MGB run (for 1 SNP) – Genotyping 1536 SNPs uses the same amount of DNA as 96 SNPs (but costs a lot more) 15

Proposed Small-Scale (1 to 100 SNPs) Genotyping Policy • Investigator(s) will make request to MESA Genetics Committee • Genotyping Sub-Committee is chaired by Mike Tsai, with Don Bowden and Kent Taylor as members • Genotyping Sub-Committee will: – Decide best way to perform the genotyping (where & how) – Combine requests into fewer Illumina runs in order to save DNA resource – Work with MESA Family genotyping committee • If Illumina design not possible, then Taq. Man MGB is available. Check feasibility at db. SNP • If these designs are not possible, then MESA family genetics committee will determine the best way to genotype the particular SNP. Emphasis will be placed on conservation of DNA 16

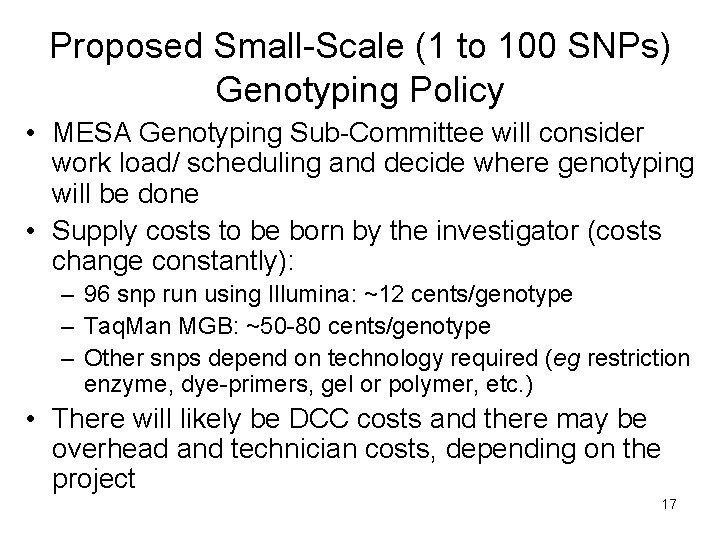
Proposed Small-Scale (1 to 100 SNPs) Genotyping Policy • MESA Genotyping Sub-Committee will consider work load/ scheduling and decide where genotyping will be done • Supply costs to be born by the investigator (costs change constantly): – 96 snp run using Illumina: ~12 cents/genotype – Taq. Man MGB: ~50 -80 cents/genotype – Other snps depend on technology required (eg restriction enzyme, dye-primers, gel or polymer, etc. ) • There will likely be DCC costs and there may be overhead and technician costs, depending on the project 17

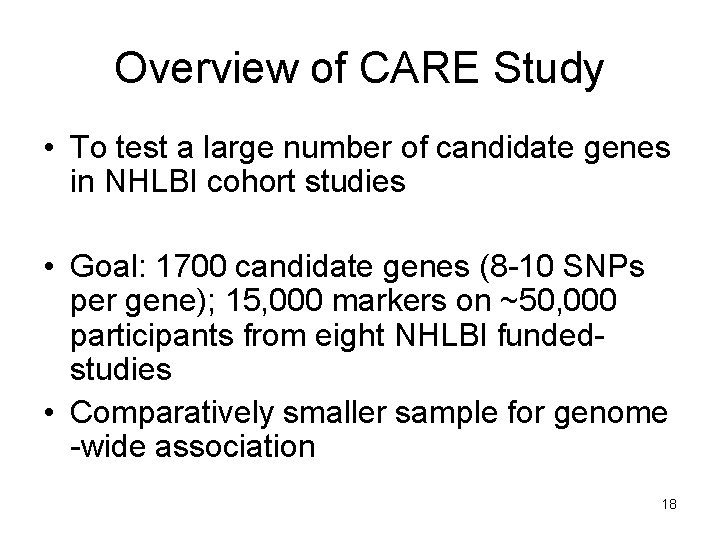
Overview of CARE Study • To test a large number of candidate genes in NHLBI cohort studies • Goal: 1700 candidate genes (8 -10 SNPs per gene); 15, 000 markers on ~50, 000 participants from eight NHLBI fundedstudies • Comparatively smaller sample for genome -wide association 18

Eight NHLBI CARE Cohorts • ARIC - Atherosclerosis Risk In Communities • CARDIA - Coronary Artery Risk Development in Young Adults • CHS - Cardiovascular Health Study • CSSCD - The Cooperative Study of Sickle Cell Disease • FHS - Framingham Heart Study • JHS - Jackson Heart Study • MESA - Multi-Ethnic Study of Atherosclerosis • SHHS - Sleep Heart Health Study 19

Organization of CARE Study • RFP to establish a Genotyping/ Coordinating Center – coordination and utilization of genetic and phenotypic data – from well-characterized NHLBI cohorts • Center funded at the Broad Institute Center for Genotyping and Analysis 20

CARE Genotyping and Analysis Center Goals 1. Receipt and management of DNA samples and phenotypic information to facilitate cross study analysis 2. Design genotyping experiments with software tools integrating sample management, SNP selection, and SNP genotyping platform 3. Support genotyping experiment execution in candidate genes and a set of genome-wide SNPs 4. Manage data and develop and apply statistical methods required to identify associations between genotypes and HLB phenotypes 21

CARE Steering Committee Larry Atwood, Boston, MA FHS Eric Boerwinkle, Houston, TX ARIC Richard Fabsitz, Bethesda, MD NHLBI Myriam Fornage, Houston, TX CARDIA Stacey Gabriel, Cambridge, MA Broad Joel Hirschhorn, Cambridge, MA Broad Ronald Krauss, Oakland, CA Heart Abdullah Kutlar, August, GA Blood Deborah Meyers, Winston-Salem, NC Lung Emanual Mignot, Palo Alto, CA Sleep Dina Paltoo, Bethesda, MD NHLBI Susan Redline, Cleveland, OH SHHS Jerome Rotter, Los Angeles, CA MESA Jeanne Smith, Englewood, NJ CSSCD Russell Tracy, Colchester, VT CHS James Wilson, Jackson, MS JHS 22

• Mediawiki-based web page http: //www. broad. mit. edu/gen_analysis/care/index. php/Main_Page • Numerous conference calls • Steering Committee Meeting, July 25, 2006 – Discussion Items • • Define pilot project (phenotypes & SNPs) Establish principles of data release Discuss genotyping study design Select phenotypes to be analyzed 23

MESA CARE Working Group Susan Heckbert Bruce Psaty Craig Johnson Stephen Richard Kronmal Jerome Rotter Kiang Liu Kent Taylor Joe Mychaleckyj Russell Tracy James Pankow Michael Tsai Wendy Post 24

CARE Subcommittees & Chair Data Release/IRB - James Wilson DNA Transfer/Genotyping - Larry Atwood Analysis/Study Design - Stephen Rich Candidate Gene/SNP Selection - Myriam Fornage Phenotypes - Bruce Psaty - Susan Heckbert Informatics - Joe Mychaleckyj Publications - to be determined 25

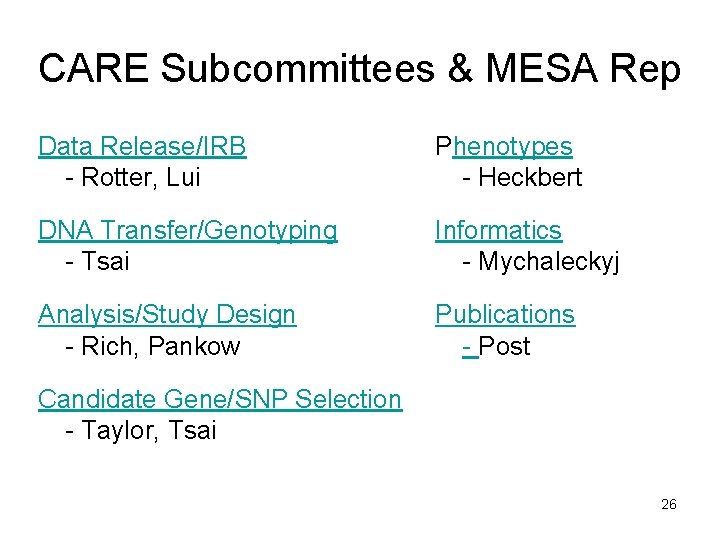
CARE Subcommittees & MESA Rep Data Release/IRB - Rotter, Lui Phenotypes - Heckbert DNA Transfer/Genotyping - Tsai Informatics - Mychaleckyj Analysis/Study Design - Rich, Pankow Publications - Post Candidate Gene/SNP Selection - Taylor, Tsai 26

General CARE Timeline • Set up infrastructure at Broad and Steering/ Subcommittees • Determine protocol for Pilot Study • Candidate Gene Study • Whole Genome Association Study 27

Current CARE Discussion Items • Pilot Study – Phenotype List – Candidate Gene List • • IRB, Data Distribution Agreement/ Policy Design/Analysis Informatics Publications Policy 28

Proposed for Pilot • • Diabetes – yes, no, pre, unknown Dyslipidemia – Total, HDL, LDL, TG Hypertension – sitting SPB and DBP, history Obesity – height, weight • Medications associated with phenotypes • How were phenotypes measured • General covariates – age, sex, race, tobacco • 22 Candidate genes (from prior literature), 1 -4 SNPs per gene 29

CARE Draft Data Distribution Policy and Data Access Agreement • NHLBI goal – – Comprehensive genotype and phenotype data set Broadly accessible to the scientific community Protect interest of study participants Promote productivity of CARE Cohort Investigators • Based on Framingham SHARE policy • Will be submitted to each Cohort’s Ancillary Studies Committee and local IRBs for approval 30

CARE Draft Data Distribution Policy and Data Access Agreement • If approval can not be reached, options include – Re-consent if funding available – Data Enclave model (investigator-driven analysis but not raw genotyping download) – Withdrawal of the study from CARE • CARE will provide final documents and IRB talking points 31

NIH RFI on Data Sharing in GWAS • NIH is requesting comments on proposed policy for sharing data obtained in NIH-supported or conducted genome-wide association studies (GWAS) • GWAS defined as “any study of genetic variation across the entire human genome that is designed to identify genetic associations with observable traits or presence or absence of a disease of condition” • Proposed policy addresses: – – Data sharing procedures Data access principles Intellectual property Protection of research participants • Comments are due Oct. 31. 32

NIH RFI on Data Sharing in GWAS • Requesting input on overall concept and specific questions: – Benefits and risks of sharing de-identified data – Additional protections to minimize risks to research participants beyond de-identification of data – Advantages and disadvantages of proposed • • Centralized repository Approach to data submission Approach to scientific publication Approach to intellectual property – Additional resources needed by investigators to meet the goals of the proposed policy 33

NIH RFI on Data Sharing in GWAS Principles • NIH believes that full value of GWAS can be realized only if the data is made available as rapidly as possible to a wide range of scientific investigators • Rapid and broad data access is important – Significant resources involved – Challenges of analyzing large datasets – Opportunities for making comparisons across multiple studies • Protection of research participants is a fundamental principle and NIH is committed to responsible stewardship of the data 34

NIH RFI on Data Sharing in GWAS Applicability • Draft policy would apply to active research applications identified by applicants or NIH staff as GWAS per Notice to Applicants for NIH Genome-Wide Association Studies (NOT-OD-06 -071, release date 05/15/06) 35

NIH RFI on Data Sharing in GWAS Data Management: Data Repository • Central GWAS data repository, NCBI-NLM • Single point of access for – Basic information on NIH-supported GWAS – Genotype-phenotype datasets • Repository will not be exclusive source of GWAS data • Repository will access GWAS datasets from other, non-NIH-supported sources 36

NIH RFI on Data Sharing in GWAS Data Management: Data Submission (1) • All investigators who receive NIH support for GWAS “are expected to submit” descriptive information about GWAS – Included in open access portion of repository • Must include – – – Protocol Questionnaires Study manuals Variables measured Other supporting documentation 37

NIH RFI on Data Sharing in GWAS Data Management: Data Submission (2) • NIH “strongly encourages” submission of curated and coded – Phenotype data – Exposure data – Genotype data – Pedigree data as soon as QC procedures have been completed at local institution • Data made available through controlled access process 38

NIH RFI on Data Sharing in GWAS Data Management: Data Submission (3) • To minimize risks to study participants – Data will be submitted without “identifiable information and using a random, unique code” • Keys to codes will be held by submitting institutions • Certification by submitting institution that identities will not be disclosed to repository or secondary users without appropriate approvals – Research participants should not expect return of individual research results 39

NIH RFI on Data Sharing in GWAS Data Management: Data Submission (4) • Submissions must include – Certification from IRB that submission to repository has been approved; specifically • Inclusion in repository and sharing of data is consistent with informed consent • Identification of uses specifically excluded by the informed consent – Statement from the institution that submission of data is in accord with all laws and regulations 40

NIH RFI on Data Sharing in GWAS Data Management: Data Access (1) • NIH Data Access Committee (DAC) – Regulates access to • Genotype-phenotype datasets • Pre-computed analyses – simple genotype-phenotype associations – Variants in LD with variants showing significant association with a phenotype or trait – DACs may be established based on programmatic areas of interest/expertise – All DACs will operate under common principles for transparency 41

NIH RFI on Data Sharing in GWAS Data Management: Data Access (2) • Data Use Certificate from investigators seeking data must include stipulations – Data will be used only for approved research – Researcher will protect data confidentiality – Applicable laws, local institutional policies and procedures will be followed – No attempt to identify participants will be made – Dataset will not be sold or shared with third parties – Researcher will provide annual progress report • Certificate must be Co-signed by Institution • Certificate will be reviewed/approved by DAC 42

NIH RFI on Data Sharing in GWAS Publications • For a “defined period of time” following release of a given dataset, submitting investigators should retain exclusive right to publish – NIH can grant access to others during this period, but they are expected not to publish – Period of exclusivity proposed at 9 months although shorter may be requested by NIH – Following expiration of exclusivity, any investigator with approved access to the data can publish • Contributing investigators, funding sources should be acknowledged in publications 43

NIH RFI on Data Sharing in GWAS Intellectual Property • The “hope of the NIH” that associations and conclusions from the data remain unencumbered by intellectual property claims • Encourages patenting technology suitable for private investment and development of products that address public needs • Filing of patent applications or enforcement of patents “could substantially diminish the utilization of information and the potential public benefit they could provide” 44

NIH RFI on Data Sharing in GWAS Summary • Investigators funded for GWAS will be expected to – Provide descriptive information about their studies • Investigators submitting data will be expected to submit – De-identified genotype, phenotype, covariate data – Limitations to use of data based on consent – Certification from IRB, Institutional assurances • Investigators requesting data will be expected to submit – Description of proposed project – Data Use Certification (protection of data confidentiality) – Annual progress reports 45

NIH RFI on Data Sharing Concerns (1) • Does this apply prospectively or retroactively? • If retroactive, current MESA consent is not consistent with proposed policy 46

NIH RFI on Data Sharing Concerns (2) • Extensive covariate data makes a subject potentially identifiable 47

NIH RFI on Data Sharing Concerns (3) • “Forensic-equivalent” genome-wide data makes a subject and their relatives potentially identifiable 48

NIH RFI on Data Sharing Concerns (4) • Consent to include genetic, phenotypic, and covariate data in a government controlled database might be challenging – Particularly in minority groups that have been previously abused by research 49

NIH RFI on Data Sharing Concerns (5) • Monitoring of data security delegated to investigators and IRBs with no direct interest in study participants 50

NIH RFI on Data Sharing Concerns (6) • It is likely that none of these cohort studies and/or their IRBs and/or their investigators and/or their subjects contemplated any type of “forensicequivalent”, wide-ranging data sharing and the accompanying potential for loss of confidentiality 51

NIH RFI on Data Sharing Summary of Concerns • Does this apply prospectively or retroactively? • If retroactive, current MESA consent is not consistent with proposed policy • Extensive covariate data makes a subject potentially identifiable • “Forensic-equivalent” genome-wide data makes a subject and their relatives potentially identifiable • Consent to include genetic, phenotypic, and covariate data in a government controlled database might be challenging – Particularly in minority groups that have been previously abused by research • Monitoring of data security delegated to investigators and IRBs with no direct interest in study participants • Question if local IRB’s of existing cohorts will approve 52
 Packing by jerome k jerome
Packing by jerome k jerome Packing by jerome k jerome
Packing by jerome k jerome No participes de la mesa de los demonios
No participes de la mesa de los demonios Rotter defined maladaptive behavior as behavior that
Rotter defined maladaptive behavior as behavior that Teori walter mischel
Teori walter mischel Perluasan perasaan diri
Perluasan perasaan diri Root rotter
Root rotter Segno di rotter
Segno di rotter Cümle tamamlama
Cümle tamamlama Cümle tamamlama
Cümle tamamlama Kelenjar rotter
Kelenjar rotter Jerry rotter
Jerry rotter Rotter cümle tamamlama testi puanlama
Rotter cümle tamamlama testi puanlama Social learning theory julian rotter
Social learning theory julian rotter Venous drainage of the upper limb
Venous drainage of the upper limb Kepribadian menurut bandura
Kepribadian menurut bandura Yashpal committee report 1993
Yashpal committee report 1993 Vaidyanathan committee
Vaidyanathan committee J j irani committee report
J j irani committee report Cadbury report ppt
Cadbury report ppt El aprendizaje por descubrimiento de jerome bruner
El aprendizaje por descubrimiento de jerome bruner Jerome kevin and seth shared a submarine sandwich
Jerome kevin and seth shared a submarine sandwich Bruner bilişsel gelişim kuramı
Bruner bilişsel gelişim kuramı Nimds
Nimds Liquidity market risk
Liquidity market risk Jerome bruner andamiaje
Jerome bruner andamiaje Jerome and hani get up early
Jerome and hani get up early Jerome bruner 3 modes of representation
Jerome bruner 3 modes of representation Piaget vygotsky bruner
Piaget vygotsky bruner Jerome and claire are doing the period of a pendulum
Jerome and claire are doing the period of a pendulum Jerome morrissey
Jerome morrissey St jerome largo fl
St jerome largo fl David jerome salinger
David jerome salinger Bruner theory of cognitive development
Bruner theory of cognitive development Jerome believes that his 4-year-old grandson
Jerome believes that his 4-year-old grandson Icfara
Icfara Jerome poivey
Jerome poivey Jerome mounsey
Jerome mounsey Cone of experience
Cone of experience Jerome bruner spouse
Jerome bruner spouse Bruner piaget and vygotsky
Bruner piaget and vygotsky Metodo singapur que es el sentido numerico
Metodo singapur que es el sentido numerico Jerome white ap calculus
Jerome white ap calculus Expresión oral según bruner
Expresión oral según bruner Ppsex
Ppsex Jerome chisholm
Jerome chisholm The night i met einstein ppt
The night i met einstein ppt Jerome freedman
Jerome freedman Assertiveness coaching
Assertiveness coaching Jerome bastin
Jerome bastin During the powerhouse lab jerome
During the powerhouse lab jerome Philippe de champaigne vanitas
Philippe de champaigne vanitas John langdon down
John langdon down