Lecture 31 DNA SYNTHESIS Mukund Modak Ph D









- Slides: 28

Lecture 31 DNA SYNTHESIS Mukund Modak, Ph. D.

DNA Objectives: • Define a DNA polymerase reaction and its components. • Where does DNA replication begin? Where does it terminate? • Know about leading and lagging strand synthesis. • How many primers are required for each strand synthesis? • What are Okazaki fragments? Remember some of the components required in lagging strand synthesis such as Pol I to remove RNA primers and DNA ligase to join the Okazaki fragments. • Know the polarity of DNA to be replicated. • Know the major eukaryotic DNA polymerases. • Remember clamp protein( PCNA in eukaryotes and B clamp in prokaryotes) that clamps DNA pol ( mainly delta and epsilon) to template and speeds up polymerase rate. • What does enzyme telomerase do? It has RNA component with sequence complimentary to telomeric( DNA) sequences.

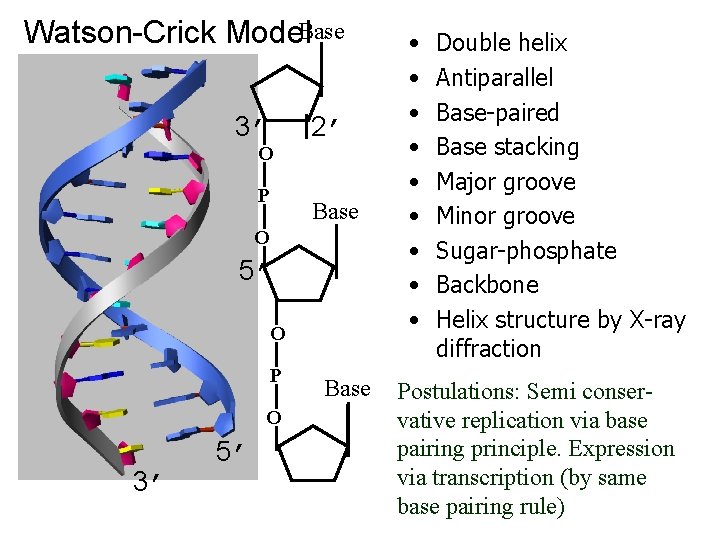
Watson-Crick Model. Base 3’ O P 2’ Base O 5’ O P O 3’ 5’ Base • • • Double helix Antiparallel Base-paired Base stacking Major groove Minor groove Sugar-phosphate Backbone Helix structure by X-ray diffraction Postulations: Semi conservative replication via base pairing principle. Expression via transcription (by same base pairing rule)

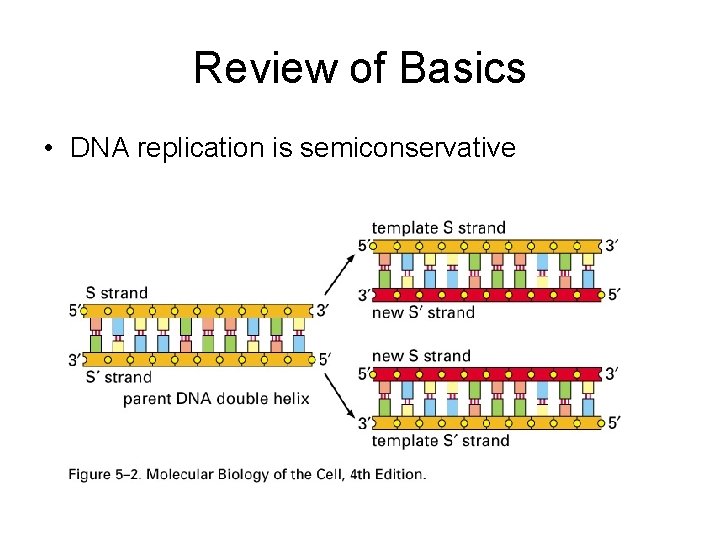
Review of Basics • DNA replication is semiconservative

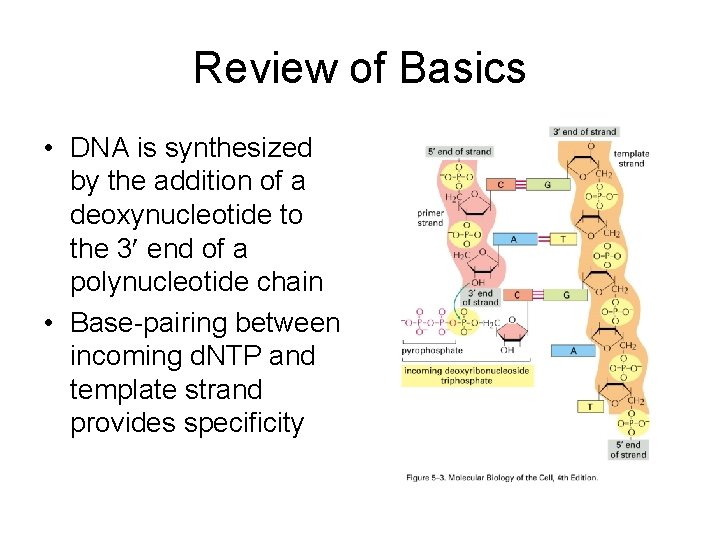
Review of Basics • DNA is synthesized by the addition of a deoxynucleotide to the 3 end of a polynucleotide chain • Base-pairing between incoming d. NTP and template strand provides specificity

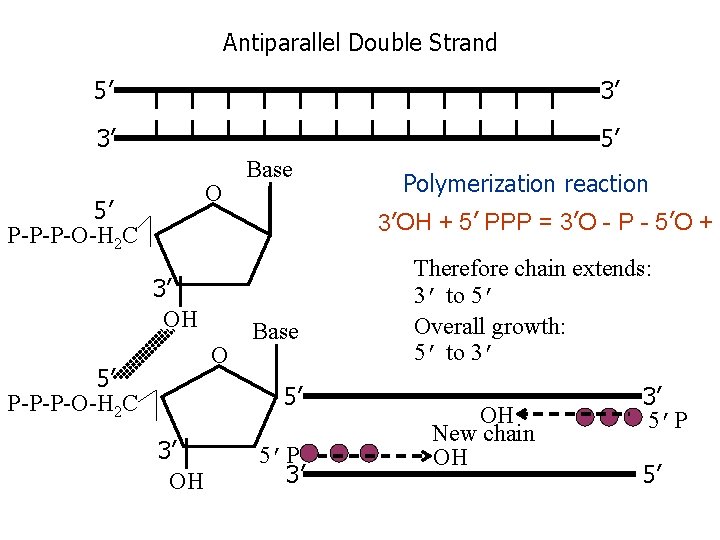
Antiparallel Double Strand 5’ 3’ 3’ 5’ O 5’ P-P-P-O-H 2 C Base Polymerization reaction 3’OH + 5’ PPP = 3’O - P - 5’O + P 3’ OH O 5’ P-P-P-O-H 2 C Base 5’ 3’ OH 5’P 3’ Therefore chain extends: 3’ to 5’ Overall growth: 5’ to 3’ OH New chain OH 3’ 5’P 5’

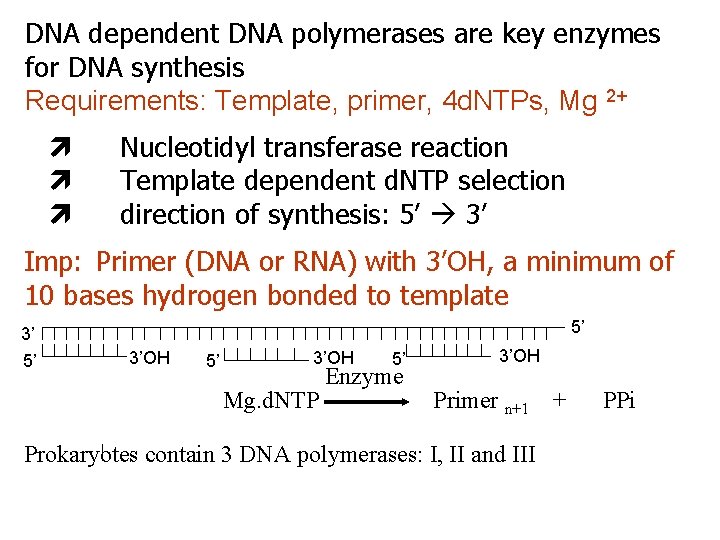
DNA dependent DNA polymerases are key enzymes for DNA synthesis Requirements: Template, primer, 4 d. NTPs, Mg 2+ Nucleotidyl transferase reaction Template dependent d. NTP selection direction of synthesis: 5’ 3’ Imp: Primer (DNA or RNA) with 3’OH, a minimum of 10 bases hydrogen bonded to template 3’ 5’ 5’ 3’OH Mg. d. NTP 5’ Enzyme 3’OH Primer n+1 Prokaryotes contain 3 DNA polymerases: I, II and III + PPi

Specialized enzymes and factors 1. Specialized polymerases that synthesize primers and DNA 2. Editing exonucleases to work with polymerases. 3. Topoisomerases that convert supercoiled DNA to relaxed form 4. Helicases that separate two parental strands of DNA 5. Accessory proteins promoting tight binding of polymerase to DNA and thereby increase the speed of polymerases (sliding clamps) Details of the process




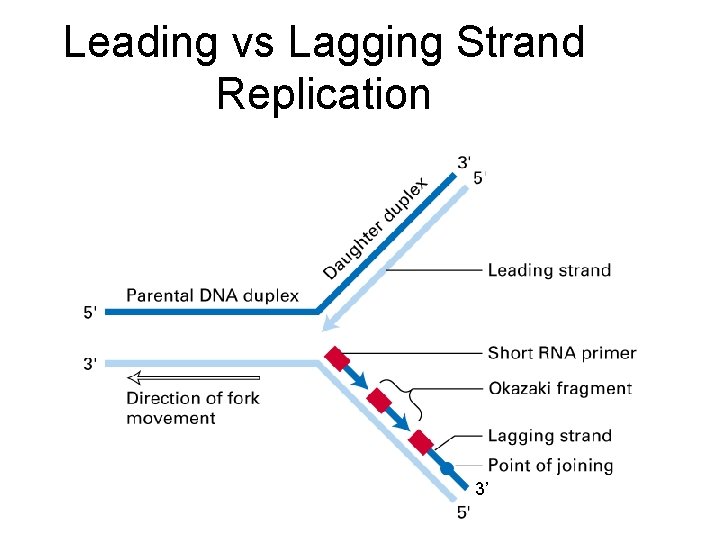
Leading vs Lagging Strand Replication 3’



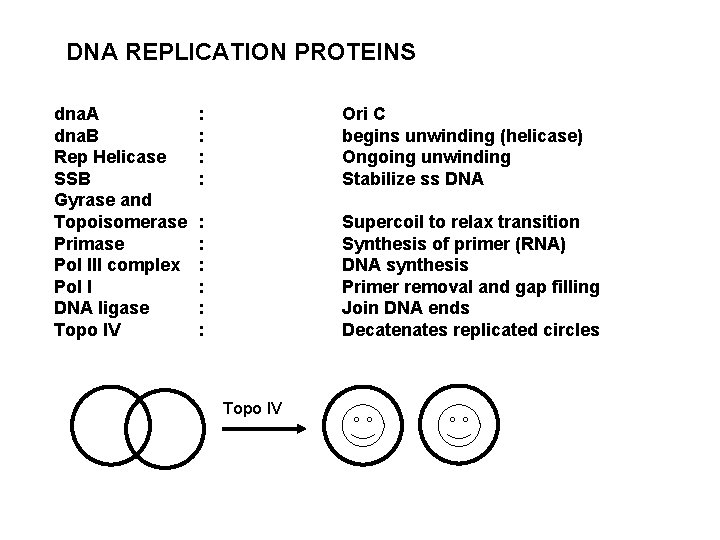
DNA REPLICATION PROTEINS dna. A dna. B Rep Helicase SSB Gyrase and Topoisomerase Primase Pol III complex Pol I DNA ligase Topo IV : : Ori C begins unwinding (helicase) Ongoing unwinding Stabilize ss DNA : : : Supercoil to relax transition Synthesis of primer (RNA) DNA synthesis Primer removal and gap filling Join DNA ends Decatenates replicated circles Topo IV

REPLICATION FORK SUMMARY Begins with unwinding of two strands with opposite polarity Two replisome assemblies (Primase + Pol III complex) Leading strand (continuous synthesis) Lagging strand (discontinuous synthesis) a. b. c. d. Looping of the strand to transiently change polarity and permit primer synthesis DNA synthesis in pieces (Okazaki fragments) Pol I to remove RNA primers and replace by DNA ligase to join small DNAs into a single large molecule with ATP or NAD as a cofactor Termination signal: TUS factor Two forks reach other at mid point of the circle and are stalled by TUS factor Replication completes with generation of catenated circles. Seperation of circles by Topo IV

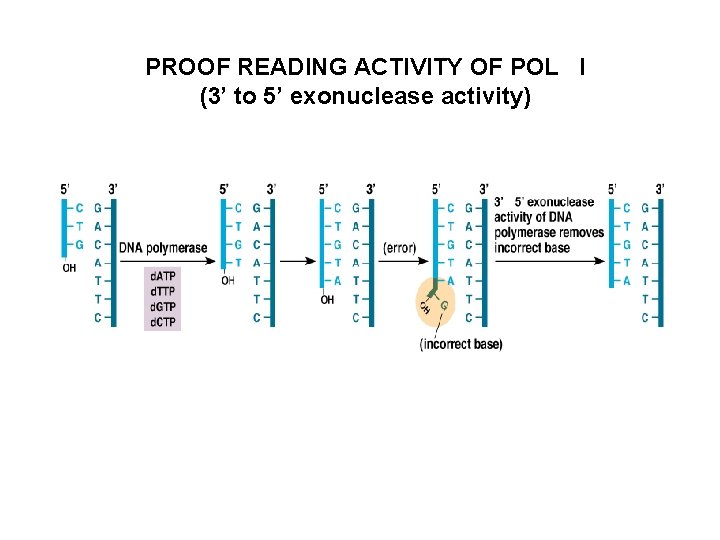
PROOF READING ACTIVITY OF POL I (3’ to 5’ exonuclease activity)

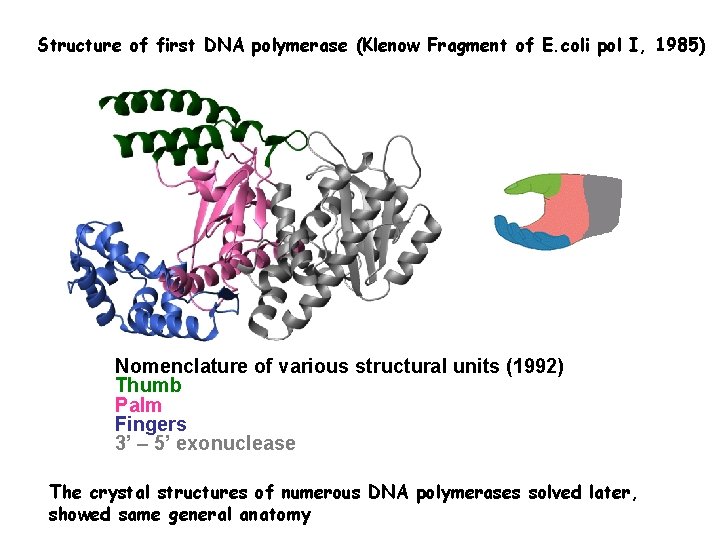
Structure of first DNA polymerase (Klenow Fragment of E. coli pol I, 1985) Nomenclature of various structural units (1992) Thumb Palm Fingers 3’ – 5’ exonuclease The crystal structures of numerous DNA polymerases solved later, showed same general anatomy

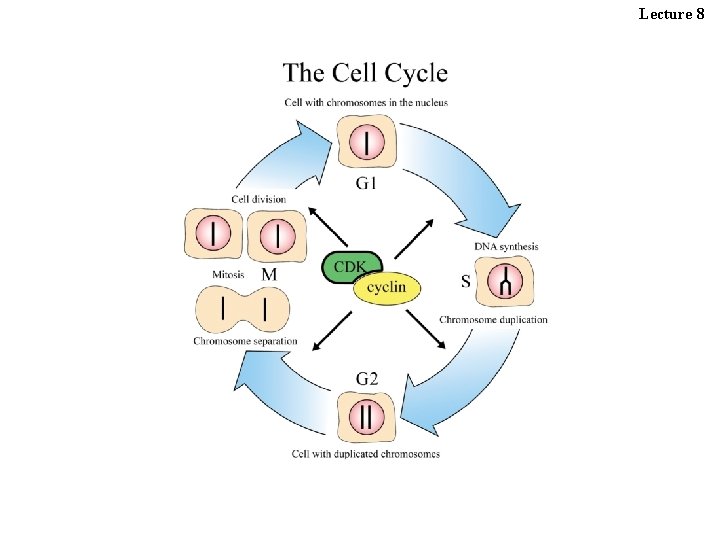
Lecture 8

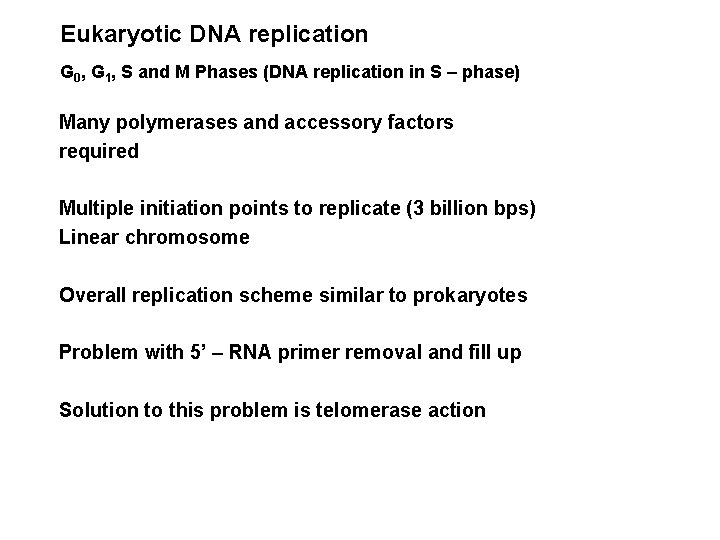
Eukaryotic DNA replication G 0, G 1, S and M Phases (DNA replication in S – phase) Many polymerases and accessory factors required Multiple initiation points to replicate (3 billion bps) Linear chromosome Overall replication scheme similar to prokaryotes Problem with 5’ – RNA primer removal and fill up Solution to this problem is telomerase action

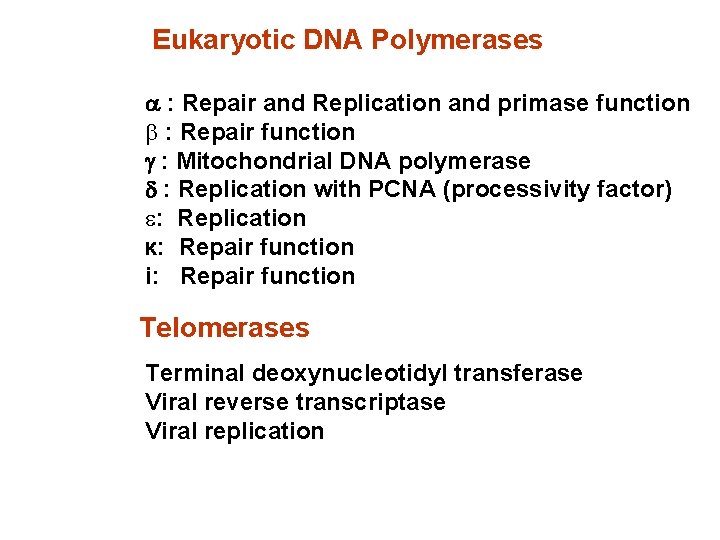
Eukaryotic DNA Polymerases a : Repair and Replication and primase function b : Repair function g : Mitochondrial DNA polymerase d : Replication with PCNA (processivity factor) : Replication к: Repair function i: Repair function Telomerases Terminal deoxynucleotidyl transferase Viral reverse transcriptase Viral replication

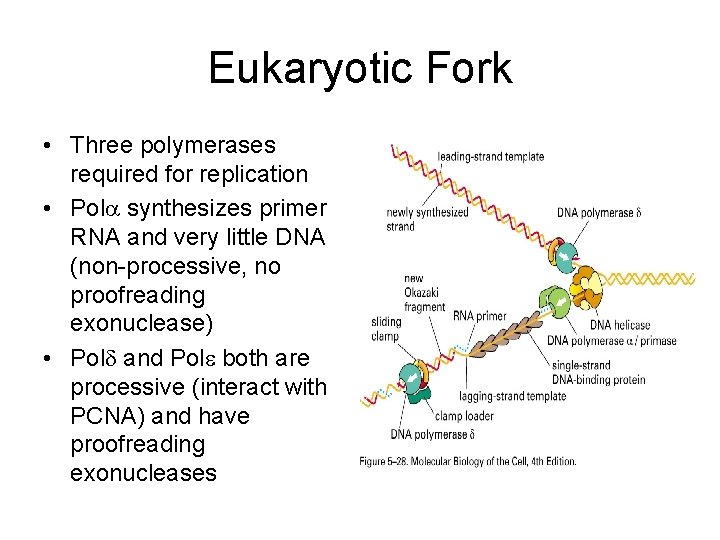
Eukaryotic Fork • Three polymerases required for replication • Pol synthesizes primer RNA and very little DNA (non-processive, no proofreading exonuclease) • Pol and Pol both are processive (interact with PCNA) and have proofreading exonucleases

Important Participants In Eukaryotic DNA Replication • Pol (alpha) (delta) and (epsilon) primer synthesis and DNA synthesis (There is a separate primase in prokaryores) • Sliding clamp or processivity factor: PCNA Increases rates and length of DNA (Equivalent to β clamp in prokaryotes) • FEN-1 nuclease removes RNA primers (In prokaryotes, 5’- nuclease activity of pol I does it) • RPA is a single strand binding protein (SSB in prokaryotes)

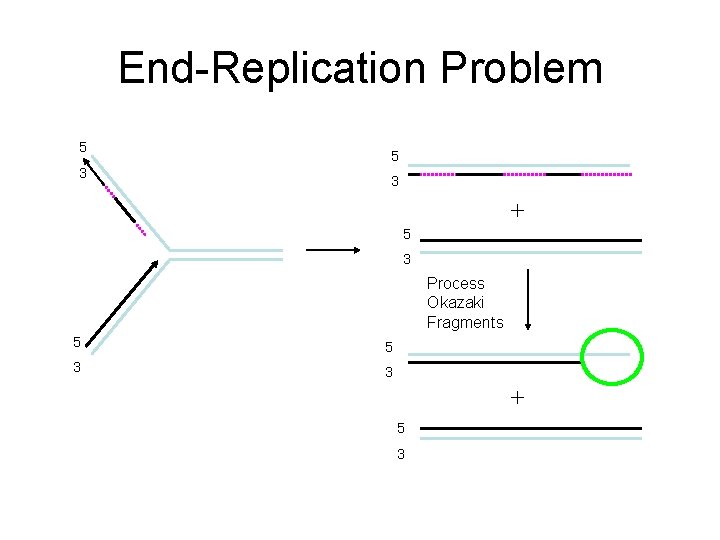
End-Replication Problem 5 3 + 5 3 Process Okazaki Fragments 5 5 3 3 + 5 3

Telomere Structure 5 3 G-rich C-rich • Telomeres composed of short (6 -10 bp) repeats (2000 – 3000) • G-rich in one strand, C-rich in other • TTAGGG/CCCTAA


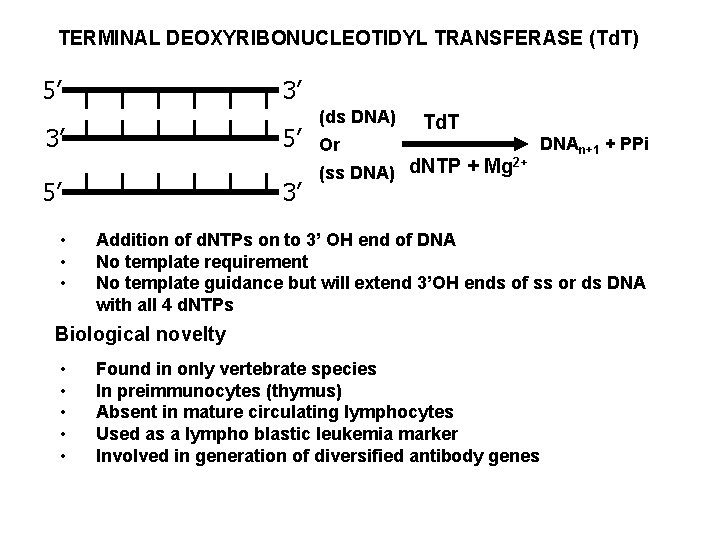
TERMINAL DEOXYRIBONUCLEOTIDYL TRANSFERASE (Td. T) 5’ 3’ 3’ 5’ 5’ • • • 3’ (ds DNA) Td. T Or 2+ (ss DNA) d. NTP + Mg DNAn+1 + PPi Addition of d. NTPs on to 3’ OH end of DNA No template requirement No template guidance but will extend 3’OH ends of ss or ds DNA with all 4 d. NTPs Biological novelty • • • Found in only vertebrate species In preimmunocytes (thymus) Absent in mature circulating lymphocytes Used as a lympho blastic leukemia marker Involved in generation of diversified antibody genes

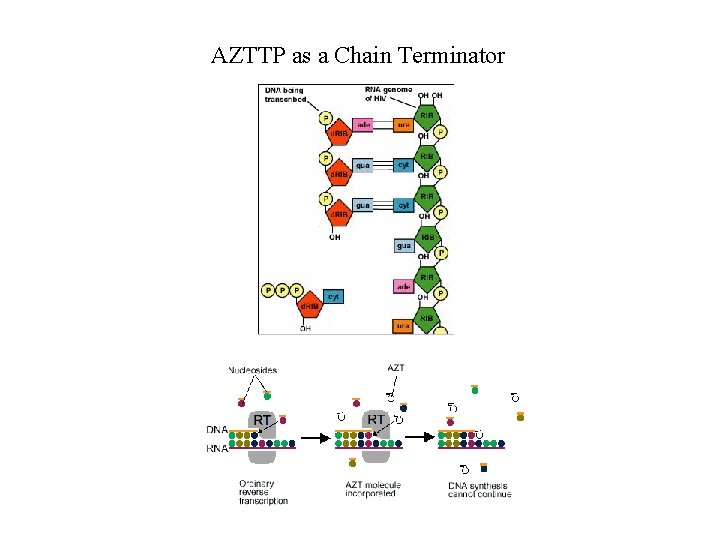
AZTTP as a Chain Terminator