Inverse Kinematics and Protein Loop Closure Presenter Chittaranjan






- Slides: 29

Inverse Kinematics and Protein Loop Closure Presenter: Chittaranjan Tripathy February 21, 2008 Figures are taken from the references unless otherwise stated. Special thanks to John Mac. Master for allowing me to use some of his slides and figures. 1

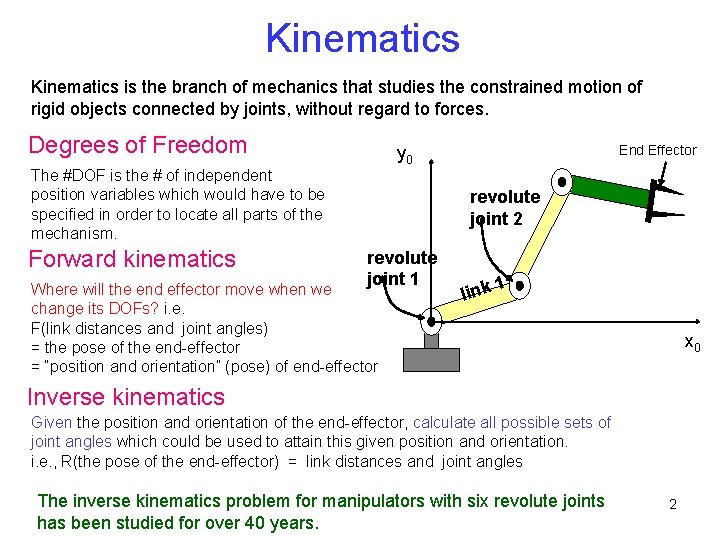
Kinematics is the branch of mechanics that studies the constrained motion of rigid objects connected by joints, without regard to forces. Degrees of Freedom The #DOF is the # of independent position variables which would have to be specified in order to locate all parts of the mechanism. Forward kinematics End Effector y 0 revolute joint 2 revolute joint 1 Where will the end effector move when we change its DOFs? i. e. F(link distances and joint angles) = the pose of the end-effector = “position and orientation” (pose) of end-effector link 1 x 0 Inverse kinematics Given the position and orientation of the end-effector, calculate all possible sets of joint angles which could be used to attain this given position and orientation. i. e. , R(the pose of the end-effector) = link distances and joint angles The inverse kinematics problem for manipulators with six revolute joints has been studied for over 40 years. 2

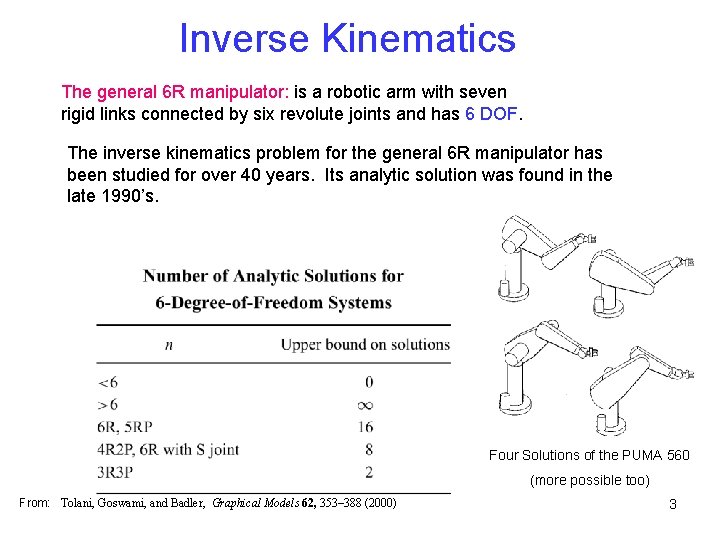
Inverse Kinematics The general 6 R manipulator: is a robotic arm with seven rigid links connected by six revolute joints and has 6 DOF. The inverse kinematics problem for the general 6 R manipulator has been studied for over 40 years. Its analytic solution was found in the late 1990’s. Four Solutions of the PUMA 560 (more possible too) From: Tolani, Goswami, and Badler, Graphical Models 62, 353– 388 (2000) 3

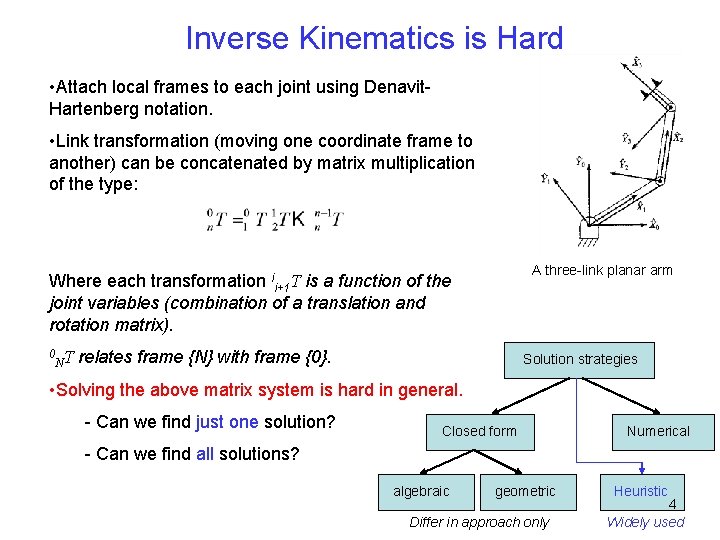
Inverse Kinematics is Hard • Attach local frames to each joint using Denavit. Hartenberg notation. • Link transformation (moving one coordinate frame to another) can be concatenated by matrix multiplication of the type: A three-link planar arm Where each transformation ii+1 T is a function of the joint variables (combination of a translation and rotation matrix). 0 NT relates frame {N} with frame {0}. Solution strategies • Solving the above matrix system is hard in general. - Can we find just one solution? Closed form Numerical - Can we find all solutions? algebraic geometric Differ in approach only Heuristic 4 Widely used

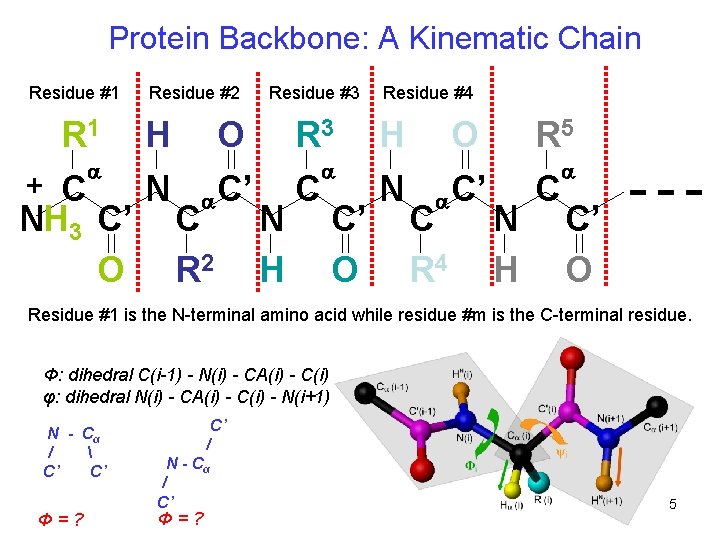
Protein Backbone: A Kinematic Chain Residue #1 R 1 + C a NH 3 C’ O Residue #2 H N Residue #3 O a C R 2 C’ R 3 N H C a C’ O Residue #4 H N O a C R 4 C’ R 5 N H C a C’ O Residue #1 is the N-terminal amino acid while residue #m is the C-terminal residue. Φ: dihedral C(i-1) - N(i) - CA(i) - C(i) φ: dihedral N(i) - CA(i) - C(i) - N(i+1) N - Cα / C’ C’ Φ=? C’ / N - Cα / C’ Φ=? 5

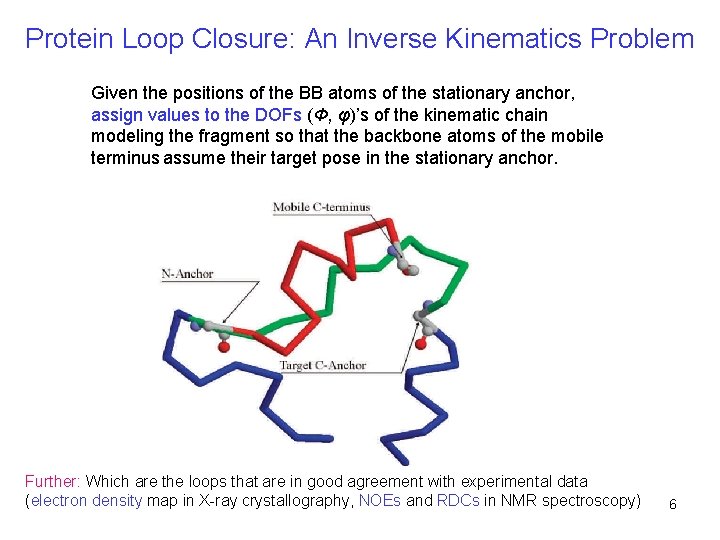
Protein Loop Closure: An Inverse Kinematics Problem Given the positions of the BB atoms of the stationary anchor, assign values to the DOFs (Φ, φ)’s of the kinematic chain modeling the fragment so that the backbone atoms of the mobile terminus assume their target pose in the stationary anchor. Further: Which are the loops that are in good agreement with experimental data (electron density map in X-ray crystallography, NOEs and RDCs in NMR spectroscopy) 6

Why Loop Closure is an Interesting Problem? Why are Loops Important? • Loops on the surface of a protein are often flexible. • Loops play important roles in binding, recognition, and active sites of proteins and enzymes. • Often difficult to characterize by X-ray crystallography as they often introduce disorder in the protein crystal. • So we see reported structures having well-defined secondary structure elements but the loops are missing! Why Loop Closure is Interesting? • IK is interesting in its own right; extensively studied in CS, Mechanical Engineering and Robotics, and in Structural biology. • Unlike Secondary structure elements in a protein, loops do not have a stereotypical structure, therefore pose a difficult challenge to compute them efficiently (with provable guarantees) from experimental data. • Often less experimental data is available for loops, and flexibility of loops may lead to larger experimental error or data that is difficult to interpret. 7

Approaches to Solve Protein Loop Closure • Ab Initio Methods • Database (Loop Library) approach. • A Greedy Heuristic: Cyclic Coordinates Descent (CCD) Algorithm • Analytical Solution to Tripeptide (6 DOF) Loop Closure. 8

Modeling Missing Loops: Ab Initio Methods • Sample from discrete set of conformational parameters, such as from the (Φ, Ψ) map, and then refine through – – Monte Carlo searches with simulated annealing. Genetic algorithms. Dynamic programming. And a few others… • Robotics inspired probabilistic sampling method. (Kavraki et. al. 1996) – Sample loop conformations ignoring constraints and later enforce the constraints through gradient descent. – Need to solve an inverse kinematics (IK) problem. 9

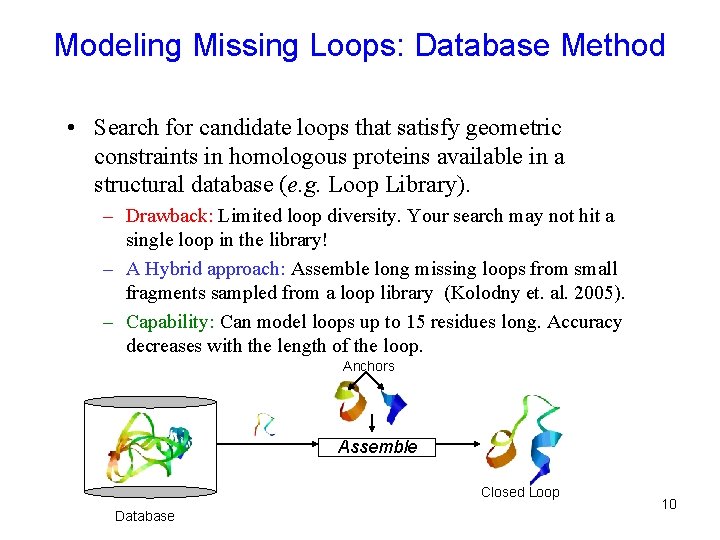
Modeling Missing Loops: Database Method • Search for candidate loops that satisfy geometric constraints in homologous proteins available in a structural database (e. g. Loop Library). – Drawback: Limited loop diversity. Your search may not hit a single loop in the library! – A Hybrid approach: Assemble long missing loops from small fragments sampled from a loop library (Kolodny et. al. 2005). – Capability: Can model loops up to 15 residues long. Accuracy decreases with the length of the loop. Anchors Assemble Closed Loop Database 10

Hybrid Approach: Ab Initio + Database Approach: • C-space is discretized by the loops/fragments in the loop library. • Represent candidate loops as a sequence of rigid building blocks (fragments) concatenated without any DOF. • Choose the fragments from the loop library (database). Issues and Technicalities: • Combinatorial Explosion due to a huge search tree. • What is an optimal length of a fragment (coarseness of sampling)? • Typically 4/5/6. How to determine the position of a new fragment? • Grow the tree from both the ends (bi-directional search). Best superimpose end three Cα atoms of the already grown chain with the first three Cα atoms of the new fragments. How to eliminate bad loops from the ensemble? - Eliminate loops that don’t agree with the experimental data. - Bad geometry, other steric and energetic parameters. 11

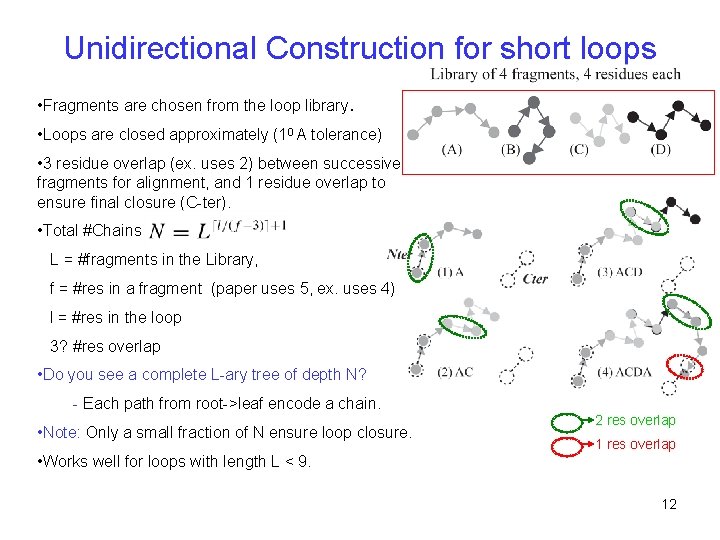
Unidirectional Construction for short loops • Fragments are chosen from the loop library. • Loops are closed approximately (10 A tolerance) • 3 residue overlap (ex. uses 2) between successive fragments for alignment, and 1 residue overlap to ensure final closure (C-ter). • Total #Chains L = #fragments in the Library, f = #res in a fragment (paper uses 5, ex. uses 4) l = #res in the loop 3? #res overlap • Do you see a complete L-ary tree of depth N? - Each path from root->leaf encode a chain. • Note: Only a small fraction of N ensure loop closure. • Works well for loops with length L < 9. 2 res overlap 12

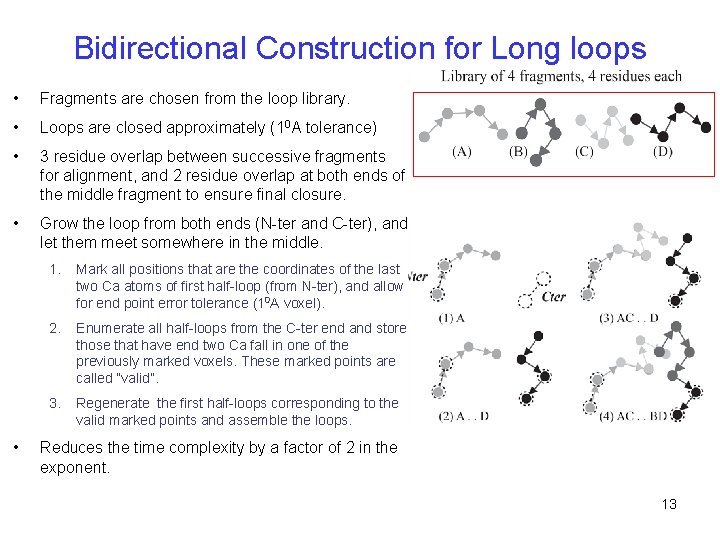
Bidirectional Construction for Long loops • Fragments are chosen from the loop library. • Loops are closed approximately (10 A tolerance) • 3 residue overlap between successive fragments for alignment, and 2 residue overlap at both ends of the middle fragment to ensure final closure. • Grow the loop from both ends (N-ter and C-ter), and let them meet somewhere in the middle. • 1. Mark all positions that are the coordinates of the last two Ca atoms of first half-loop (from N-ter), and allow for end point error tolerance (10 A voxel). 2. Enumerate all half-loops from the C-ter end and store those that have end two Ca fall in one of the previously marked voxels. These marked points are called “valid”. 3. Regenerate the first half-loops corresponding to the valid marked points and assemble the loops. Reduces the time complexity by a factor of 2 in the exponent. 13

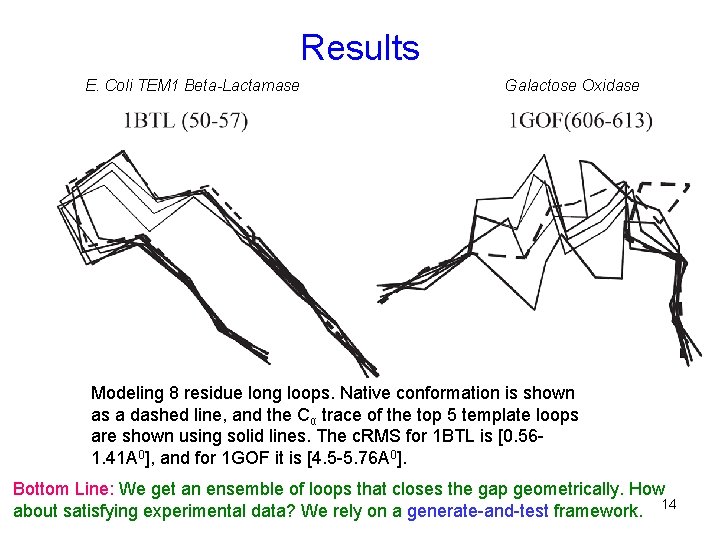
Results E. Coli TEM 1 Beta-Lactamase Galactose Oxidase Modeling 8 residue long loops. Native conformation is shown as a dashed line, and the Cα trace of the top 5 template loops are shown using solid lines. The c. RMS for 1 BTL is [0. 561. 41 A 0], and for 1 GOF it is [4. 5 -5. 76 A 0]. Bottom Line: We get an ensemble of loops that closes the gap geometrically. How about satisfying experimental data? We rely on a generate-and-test framework. 14

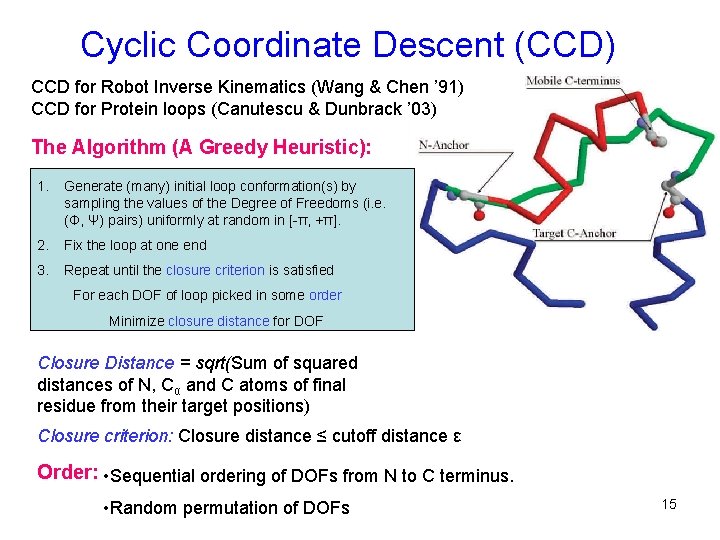
Cyclic Coordinate Descent (CCD) CCD for Robot Inverse Kinematics (Wang & Chen ’ 91) CCD for Protein loops (Canutescu & Dunbrack ’ 03) The Algorithm (A Greedy Heuristic): 1. Generate (many) initial loop conformation(s) by sampling the values of the Degree of Freedoms (i. e. (Φ, Ψ) pairs) uniformly at random in [-π, +π]. 2. Fix the loop at one end 3. Repeat until the closure criterion is satisfied For each DOF of loop picked in some order Minimize closure distance for DOF Closure Distance = sqrt(Sum of squared distances of N, Cα and C atoms of final residue from their target positions) Closure criterion: Closure distance ≤ cutoff distance ε Order: • Sequential ordering of DOFs from N to C terminus. • Random permutation of DOFs 15

Cyclic Coordinate Descent… Why CCD? • Simple to implement and extremely fast! • CCD algorithm is an optimization based algorithm to solve IK problems. Here the problem is recast as a minimization problem. • Numerically stable. • (Some) External constraints on DOFs can be integrated with predictable behavior. • Linear time complexity in the number of DOFs. Drawbacks: • It is not guaranteed to return all solutions. • It may miss out a solution even if it exists. • Some of the initial fragments don’t even close while doing a CCD on them. • Not friendly towards integrating additional constraints from certain type of 16 experimental data.

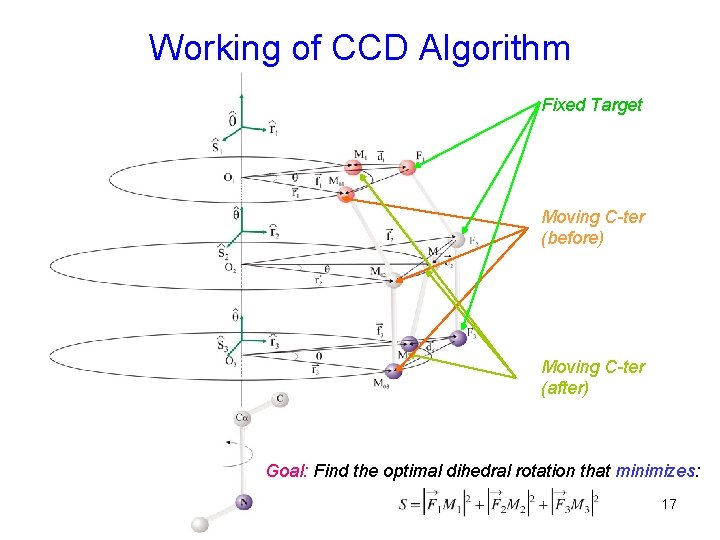
Working of CCD Algorithm Fixed Target Moving C-ter (before) Moving C-ter (after) Goal: Find the optimal dihedral rotation that minimizes: 17

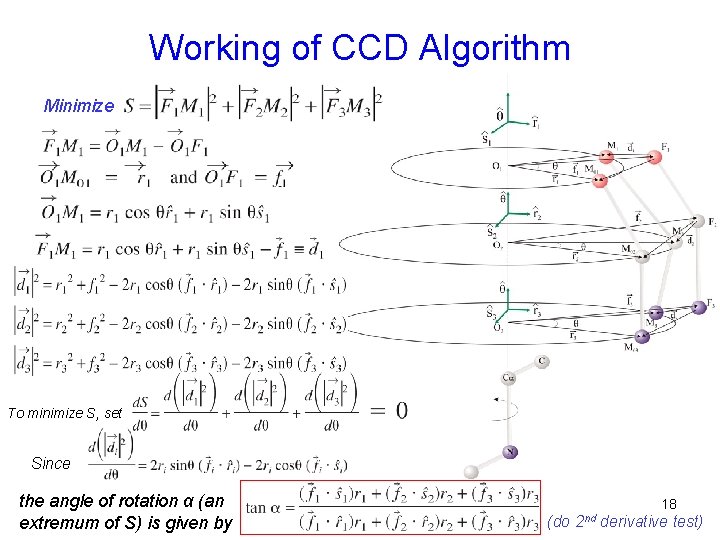
Working of CCD Algorithm Minimize To minimize S, set Since the angle of rotation α (an extremum of S) is given by 18 (do 2 nd derivative test)

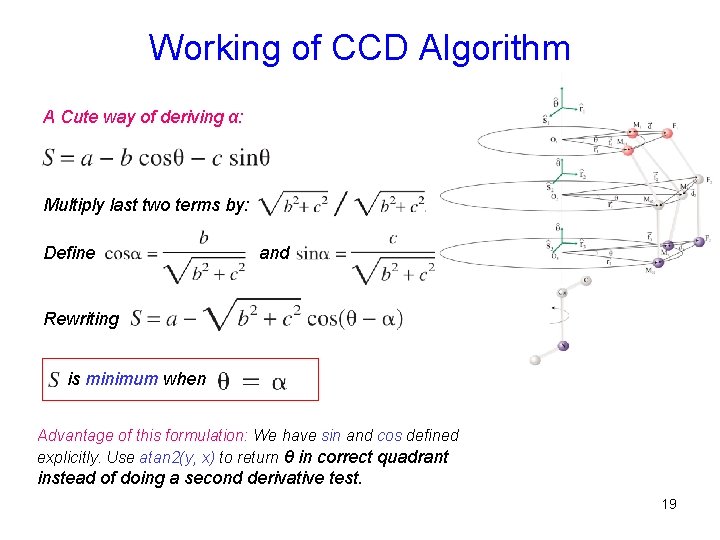
Working of CCD Algorithm A Cute way of deriving α: Multiply last two terms by: Define and Rewriting is minimum when Advantage of this formulation: We have sin and cos defined explicitly. Use atan 2(y, x) to return θ in correct quadrant instead of doing a second derivative test. 19

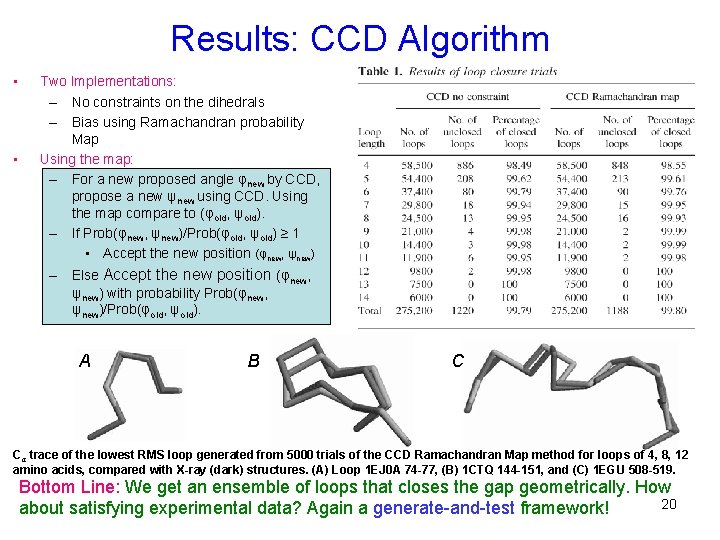
Results: CCD Algorithm • • Two Implementations: – No constraints on the dihedrals – Bias using Ramachandran probability Map Using the map: – For a new proposed angle φnew by CCD, propose a new ψnew using CCD. Using the map compare to (φold, ψold). – If Prob(φnew, ψnew)/Prob(φold, ψold) ≥ 1 • Accept the new position (φnew, ψnew) – Else Accept the new position (φnew, ψnew) with probability Prob(φnew, ψnew)/Prob(φold, ψold). A B C Cα trace of the lowest RMS loop generated from 5000 trials of the CCD Ramachandran Map method for loops of 4, 8, 12 amino acids, compared with X-ray (dark) structures. (A) Loop 1 EJ 0 A 74 -77, (B) 1 CTQ 144 -151, and (C) 1 EGU 508 -519. Bottom Line: We get an ensemble of loops that closes the gap geometrically. How 20 about satisfying experimental data? Again a generate-and-test framework!

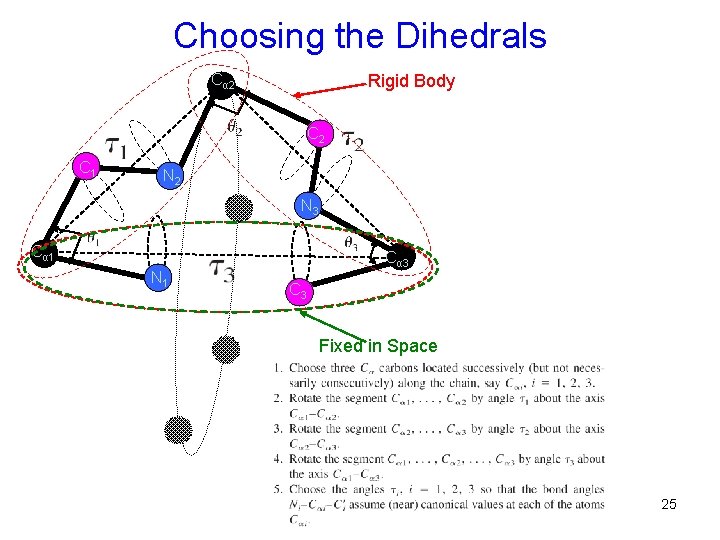
Tri-Peptide Loop Closure The Problem: finding the ensemble of possible backbone structures of a chain segment (with 6 DOFs) of a protein molecule that is geometrically consistent with preceding and following parts of the chain whose structures are given. What is Achieved here: • Solve tri-peptide loop closure (six-torsion loop closure) analytically. Tri-peptide loop closure 16 -deg polynomial in one variable Loop Conformations • Torsion angles need not be consecutive (intervening fragments must be rigid). • Can be useful in sampling longer loops when combined with an existing loop construction algorithm. • Can be used to implement a set of local moves for Monte Carlo minimization. 21

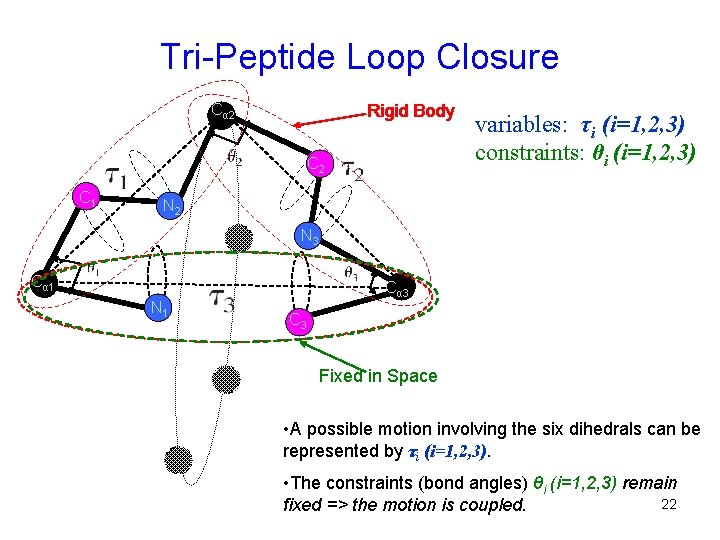
Tri-Peptide Loop Closure Cα 2 Rigid Body C 2 C 1 variables: τi (i=1, 2, 3) constraints: θi (i=1, 2, 3) N 2 N 3 Cα 1 N 1 Cα 3 C 3 Fixed in Space • A possible motion involving the six dihedrals can be represented by τi (i=1, 2, 3). • The constraints (bond angles) θi (i=1, 2, 3) remain 22 fixed => the motion is coupled.

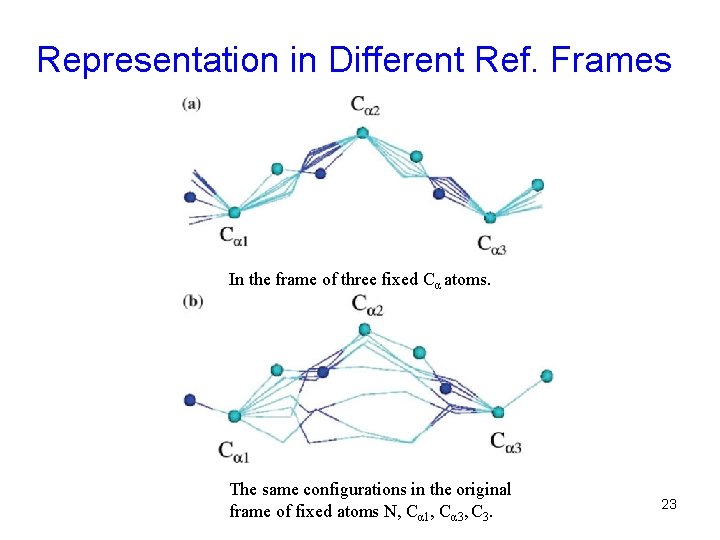
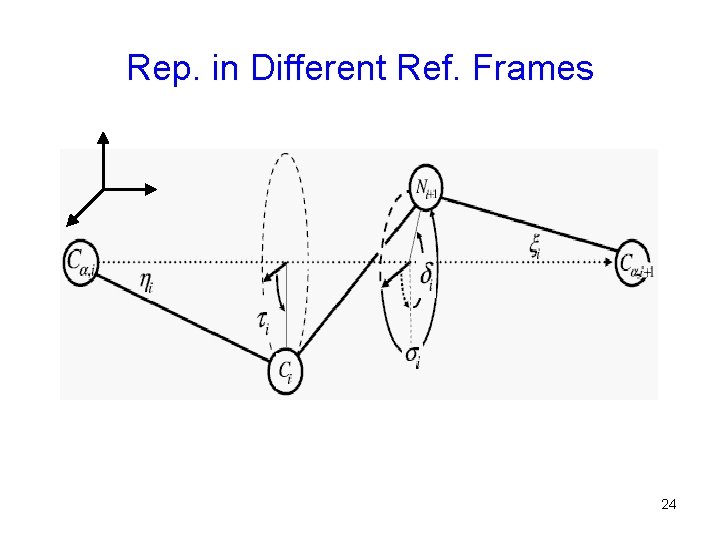
Representation in Different Ref. Frames In the frame of three fixed Cα atoms. The same configurations in the original frame of fixed atoms N, Cα 1, Cα 3, C 3. 23

Rep. in Different Ref. Frames 24

Choosing the Dihedrals Cα 2 Rigid Body C 2 C 1 N 2 N 3 Cα 1 N 1 Cα 3 C 3 Fixed in Space 25

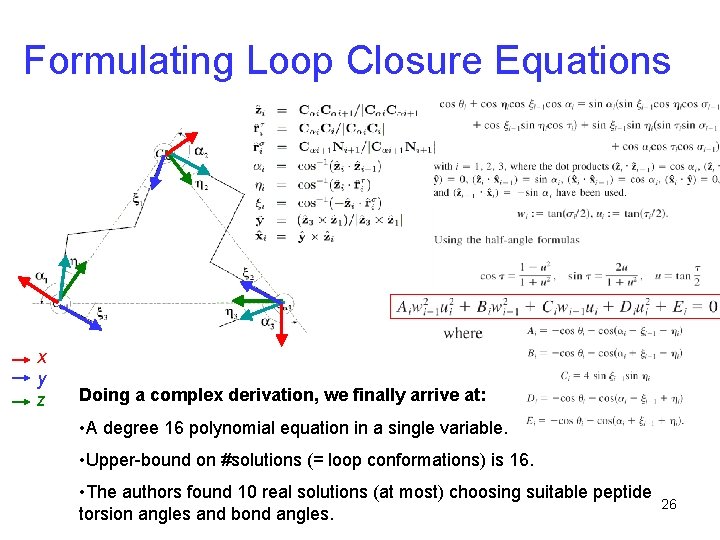
Formulating Loop Closure Equations x y z Doing a complex derivation, we finally arrive at: • A degree 16 polynomial equation in a single variable. • Upper-bound on #solutions (= loop conformations) is 16. • The authors found 10 real solutions (at most) choosing suitable peptide torsion angles and bond angles. 26

Acknowledgements Prof. Bruce Donald John Mac. Master Ed Triplett Michael Zeng 27

Thank You… 28

References 1. A. A. Canutescu and R. L. Dunbrack Jr. Cyclic coordinate descent: A robotics algorithm for protein loop closure. Protein Science, 12: 963 -972, 2003. 2. I. Z. Emiris, E. D. Fritzilas, and D. Manocha. Algebraic algorithms for determining structure in biological Chemistry. International Journal of Quantum Chemistry, Spec. Issue on Symbolic Methods, 2005. 3. E. Coutsias, C. Seok, and M. Jacobson, and K. Dill. (2004). A Kinematic View of Loop Closure. Journal of Computational Chemistry, 25, 510 -528. 4. R. Kolodny, L. Guibas, M. Levitt and P. Koehl. Inverse kinematics in biology: the protein loop closure problem. International Journal of Robotics Research, 24, 151 -163 (2005). 5. L. Wang, R. Mettu, and B. R. Donald. A Polynomial-Time Algorithm for De Novo Protein Backbone Structure Determination from NMR Data. Journal of Computational Biology, 13(7): 1276 -1288, 2006. 6. J. J. Craig. Introduction to Robotics: Mechanics and Control. 2 nd Edition, Boston, MA: Addison-Wesley 1989, 450 pp. 7. Shehu A, Clementi C, Kavraki LE. Modeling protein conformational ensembles: from missing loops to equilibrium fluctuations. Proteins. 2006 Oct 1; 65(1): 164 -179. 29