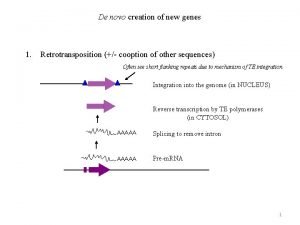
Horisontal Lateral gene transfer Genes in flux Mechanisms
- Slides: 47

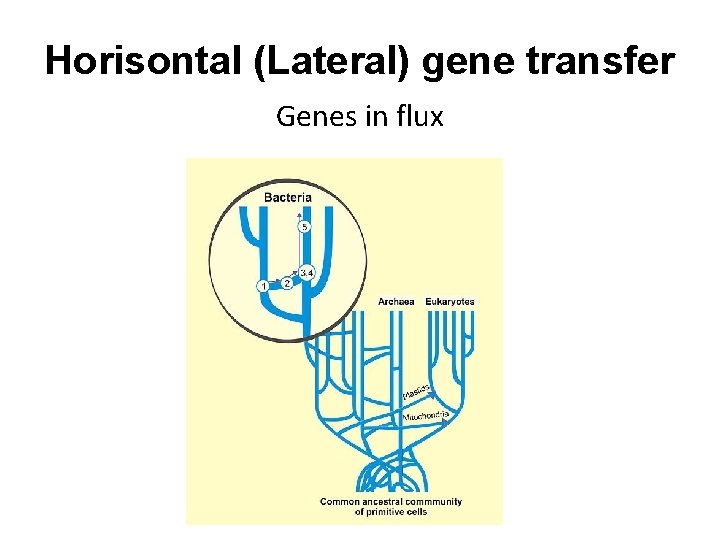
Horisontal (Lateral) gene transfer Genes in flux

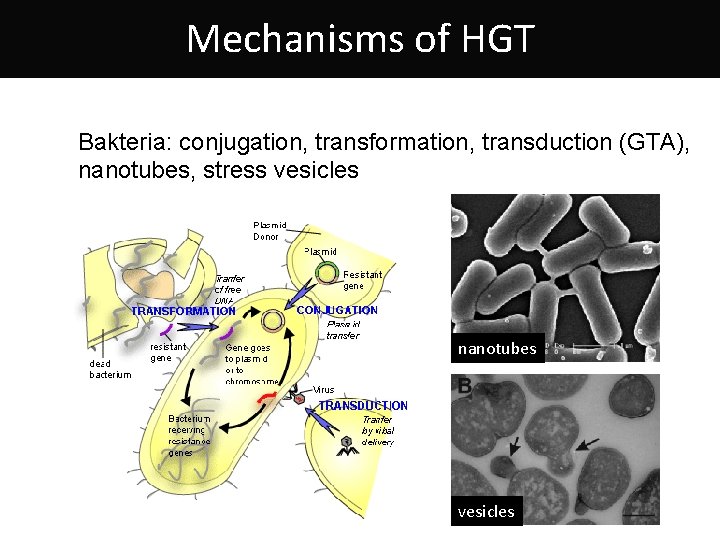
Mechanisms of HGT Bakteria: conjugation, transformation, transduction (GTA), nanotubes, stress vesicles nanotubes vesicles

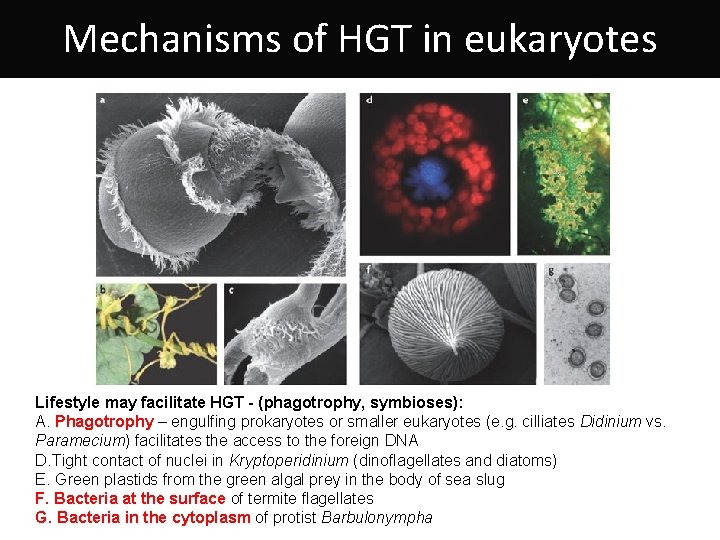
Mechanisms of HGT in eukaryotes Lifestyle may facilitate HGT - (phagotrophy, symbioses): A. Phagotrophy – engulfing prokaryotes or smaller eukaryotes (e. g. cilliates Didinium vs. Paramecium) facilitates the access to the foreign DNA D. Tight contact of nuclei in Kryptoperidinium (dinoflagellates and diatoms) E. Green plastids from the green algal prey in the body of sea slug F. Bacteria at the surface of termite flagellates G. Bacteria in the cytoplasm of protist Barbulonympha

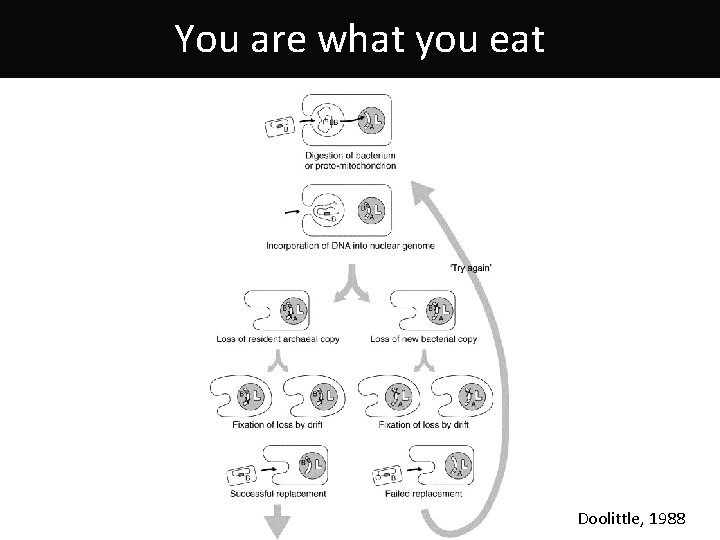
You are what you eat Doolittle, 1988

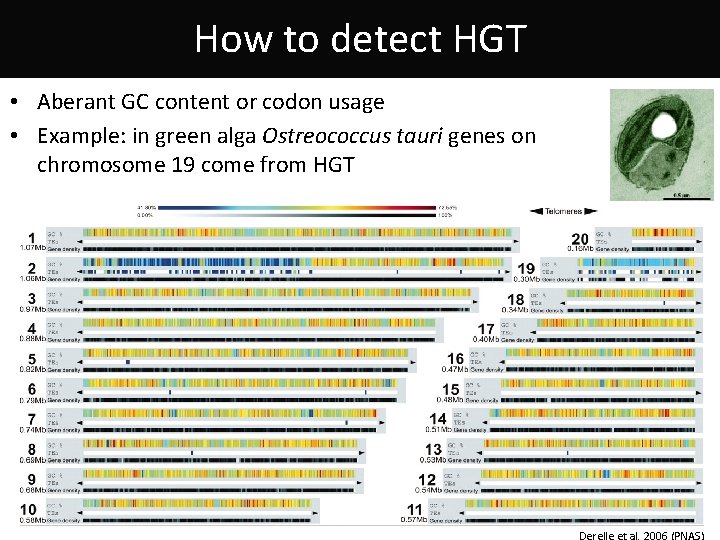
How to detect HGT • Aberant GC content or codon usage • Example: in green alga Ostreococcus tauri genes on chromosome 19 come from HGT Derelle et al. 2006 (PNAS)

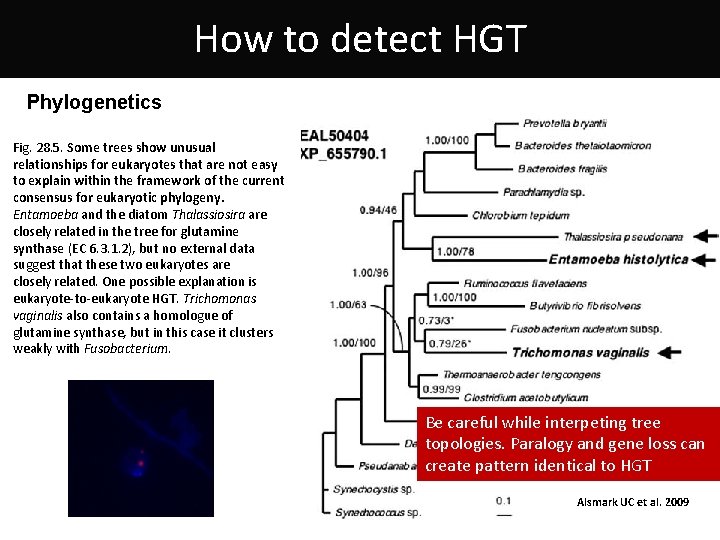
How to detect HGT Phylogenetics Fig. 28. 5. Some trees show unusual relationships for eukaryotes that are not easy to explain within the framework of the current consensus for eukaryotic phylogeny. Entamoeba and the diatom Thalassiosira are closely related in the tree for glutamine synthase (EC 6. 3. 1. 2), but no external data suggest that these two eukaryotes are closely related. One possible explanation is eukaryote-to-eukaryote HGT. Trichomonas vaginalis also contains a homologue of glutamine synthase, but in this case it clusters weakly with Fusobacterium. Be careful while interpeting tree topologies. Paralogy and gene loss can create pattern identical to HGT Alsmark UC et al. 2009

Hypothesis of complexity • HGT is more common in operational (metabolic) genes • Informational genes (transcription and translaction) are rarely affected by HGT because they have higher connectivity – are components of complex systems and interact with many proteins, form complexes. Their transfer to foreign environment is more complicated

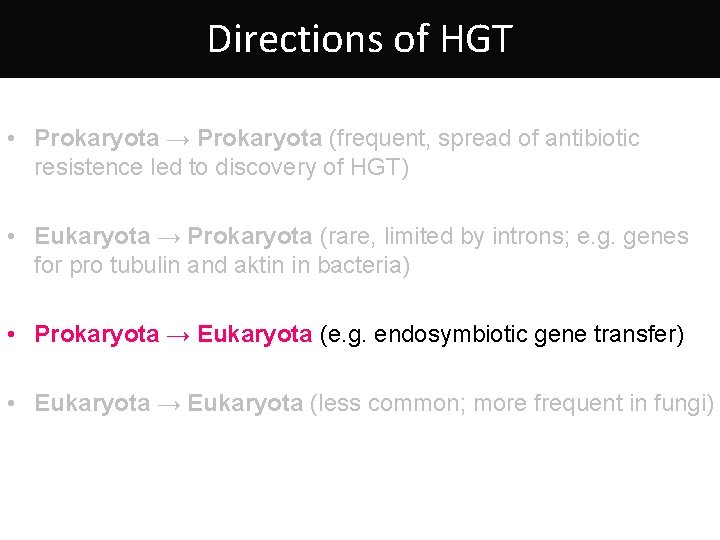
Directions of HGT • Prokaryota → Prokaryota (frequent, spread of antibiotic resistence led to discovery of HGT) • Eukaryota → Prokaryota (rare, limited by introns; e. g. genes for pro tubulin and aktin in bacteria) • Prokaryota → Eukaryota (e. g. endosymbiotic gene transfer) • Eukaryota → Eukaryota (less common; more frequent in fungi)

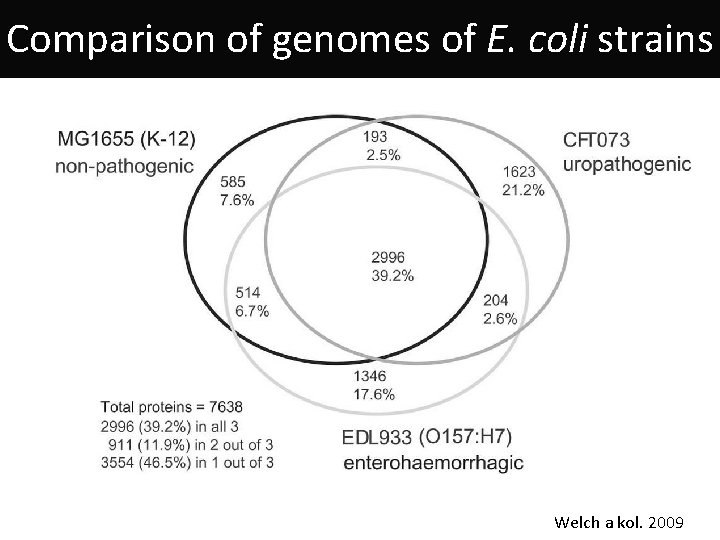
Comparison of genomes of E. coli strains Welch a kol. 2009

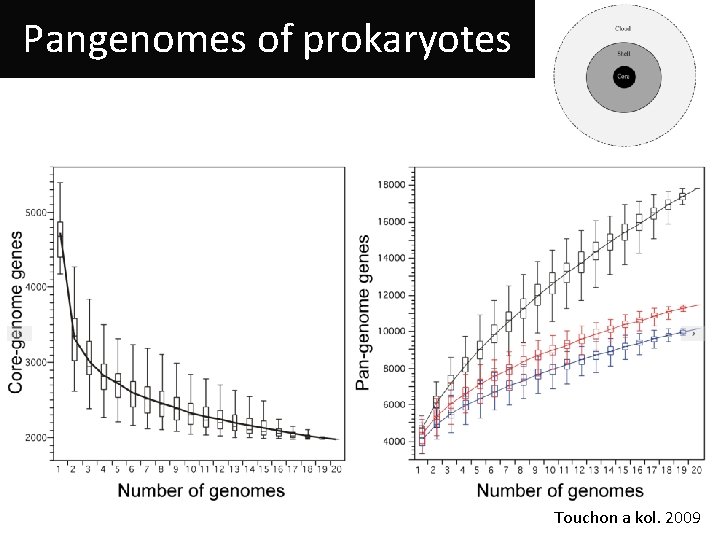
Pangenomes of prokaryotes Touchon a kol. 2009

Directions of HGT • Prokaryota → Prokaryota (frequent, spread of antibiotic resistence led to discovery of HGT) • Eukaryota → Prokaryota (rare, limited by introns; e. g. genes for pro tubulin and aktin in bacteria) • Prokaryota → Eukaryota (e. g. endosymbiotic gene transfer) • Eukaryota → Eukaryota (less common; more frequent in fungi)

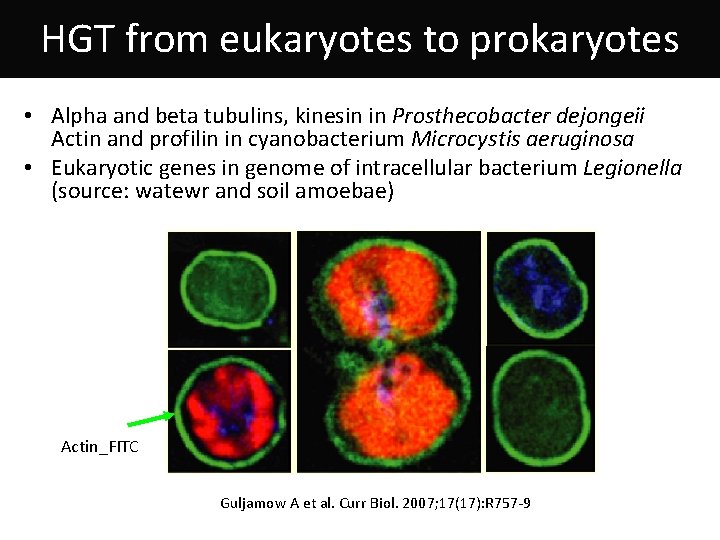
HGT from eukaryotes to prokaryotes • Alpha and beta tubulins, kinesin in Prosthecobacter dejongeii Actin and profilin in cyanobacterium Microcystis aeruginosa • Eukaryotic genes in genome of intracellular bacterium Legionella (source: watewr and soil amoebae) Actin_FITC Guljamow A et al. Curr Biol. 2007; 17(17): R 757 -9

Does a eukaryotig HGT exist? Topic of essay

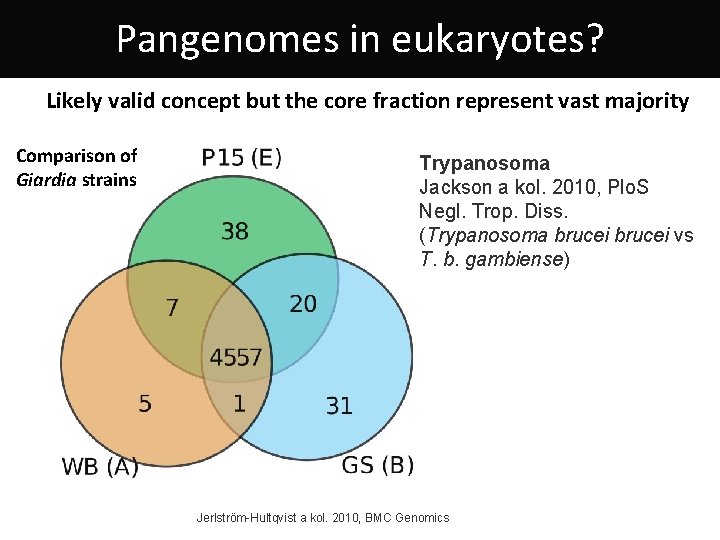
Pangenomes in eukaryotes? Likely valid concept but the core fraction represent vast majority Comparison of Giardia strains Trypanosoma Jackson a kol. 2010, Plo. S Negl. Trop. Diss. (Trypanosoma brucei vs T. b. gambiense) Jerlström-Hultqvist a kol. 2010, BMC Genomics

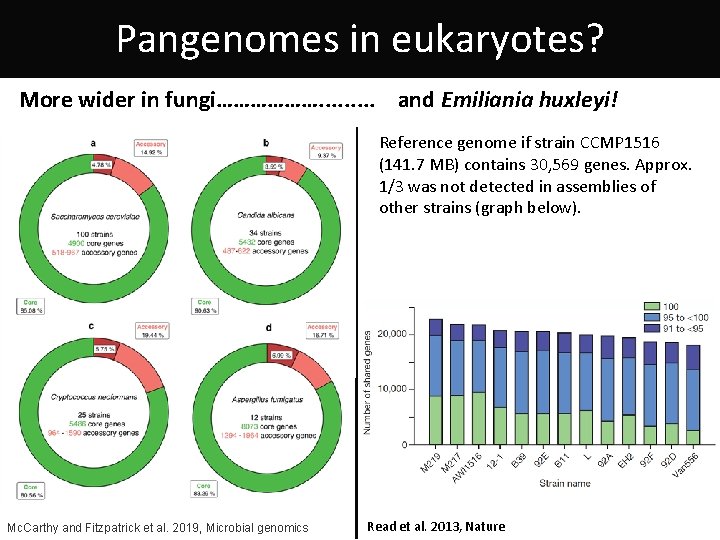
Pangenomes in eukaryotes? More wider in fungi………………. . and Emiliania huxleyi! Reference genome if strain CCMP 1516 (141. 7 MB) contains 30, 569 genes. Approx. 1/3 was not detected in assemblies of other strains (graph below). Mc. Carthy and Fitzpatrick et al. 2019, Microbial genomics Read et al. 2013, Nature

Directions of HGT • Prokaryota → Prokaryota (frequent, spread of antibiotic resistence led to discovery of HGT) • Eukaryota → Prokaryota (rare, limited by introns; e. g. genes for pro tubulin and aktin in bacteria) • Prokaryota → Eukaryota (e. g. endosymbiotic gene transfer) • Eukaryota → Eukaryota (less common; more frequent in fungi)

Adaptive function of bacterial genes • HGT is source of new adaptations • Ecologically specialised protists (anaerobic parasites, rumen ciliates, extremophiles) all have relatively number of HGT genes • But not only, marine diatom Phaedactylum tricornutum contains > 5% HGT genes. Bachorový nálevník Ophryoscolex Řasa Galdieria sulphuraria Trichomonas vaginalis

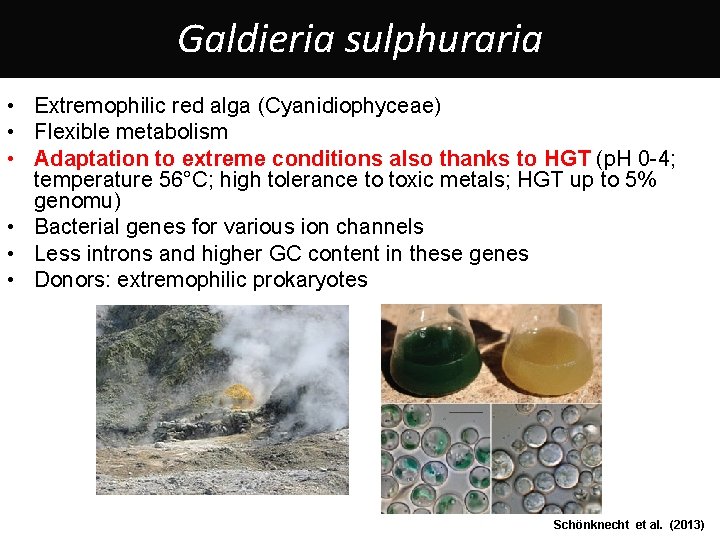
Galdieria sulphuraria • Extremophilic red alga (Cyanidiophyceae) • Flexible metabolism • Adaptation to extreme conditions also thanks to HGT (p. H 0 -4; temperature 56°C; high tolerance to toxic metals; HGT up to 5% genomu) • Bacterial genes for various ion channels • Less introns and higher GC content in these genes • Donors: extremophilic prokaryotes Schönknecht et al. (2013)

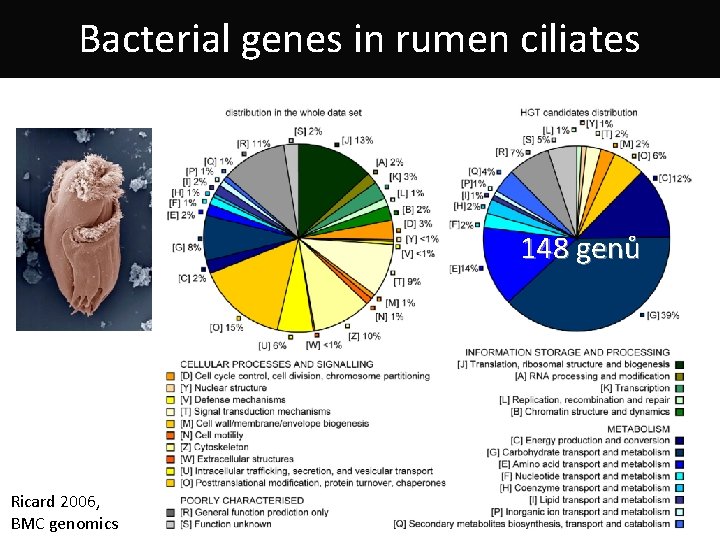
Bacterial genes in rumen ciliates 148 genů Ricard 2006, BMC genomics

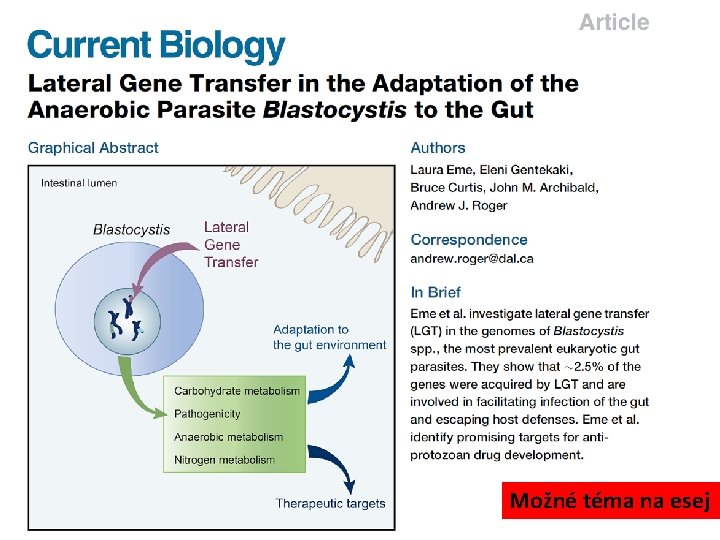
Možné téma na esej

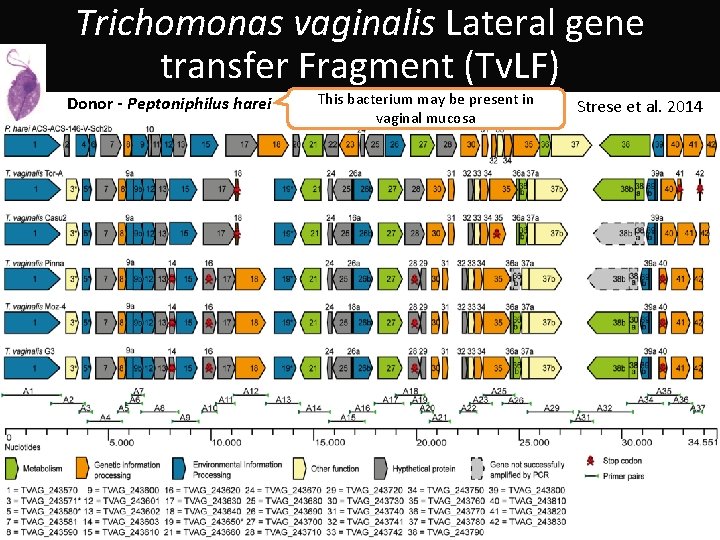
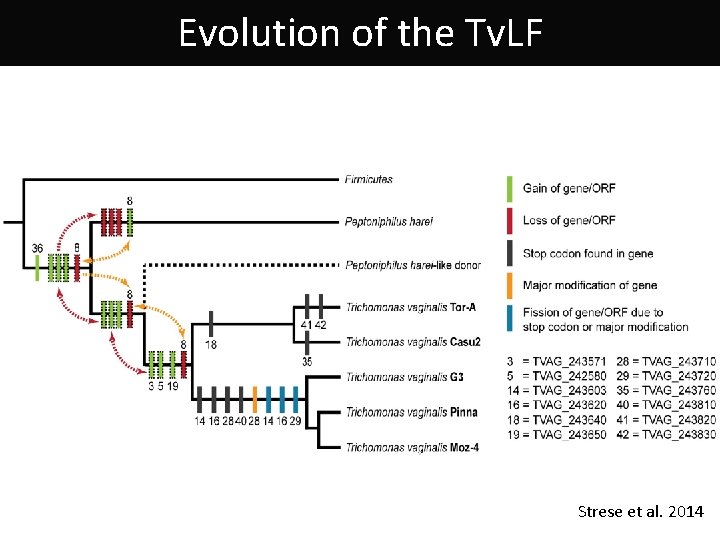
Trichomonas vaginalis Lateral gene transfer Fragment (Tv. LF) Donor - Peptoniphilus harei This bacterium may be present in vaginal mucosa Tv. LF Strese et al. 2014

Evolution of the Tv. LF Strese et al. 2014

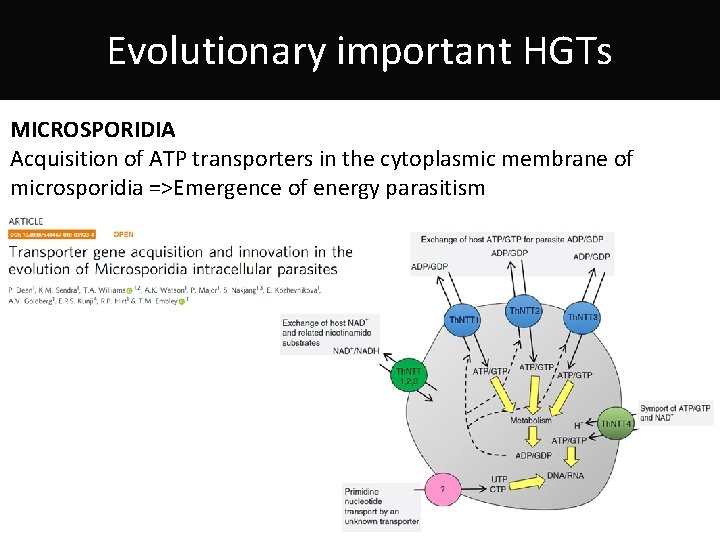
Evolutionary important HGTs MICROSPORIDIA Acquisition of ATP transporters in the cytoplasmic membrane of microsporidia =>Emergence of energy parasitism

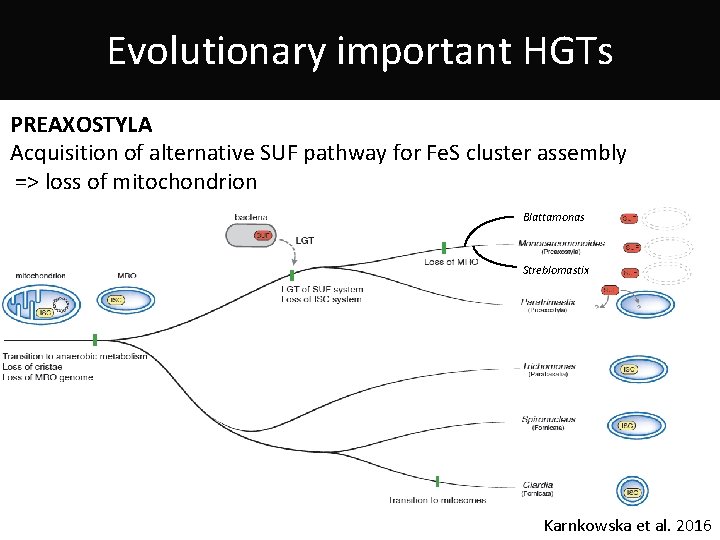
Evolutionary important HGTs PREAXOSTYLA Acquisition of alternative SUF pathway for Fe. S cluster assembly => loss of mitochondrion Blattamonas Streblomastix Karnkowska et al. 2016

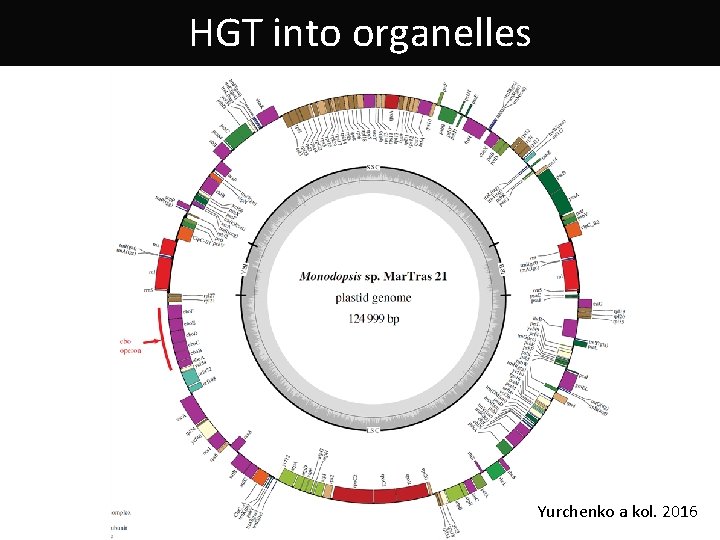
HGT into organelles Yurchenko a kol. 2016

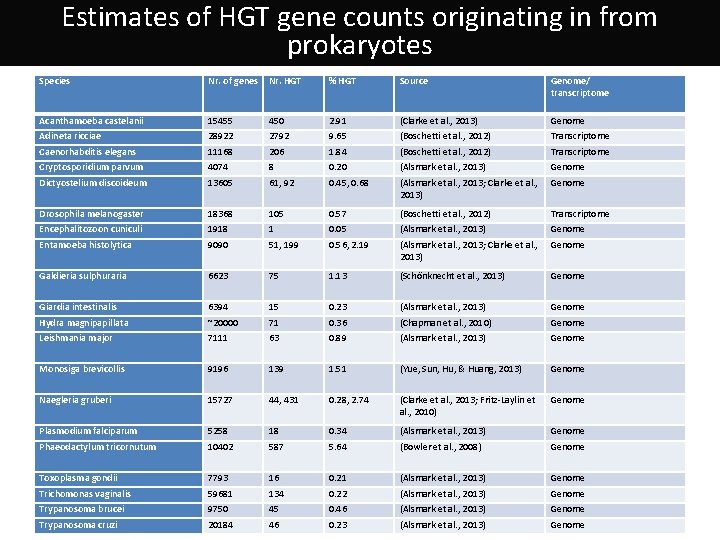
Estimates of HGT gene counts originating in from prokaryotes Species Nr. of genes Nr. HGT % HGT Source Genome/ transcriptome Acanthamoeba castelanii 15455 450 2. 91 (Clarke et al. , 2013) Genome Adineta ricciae 28922 2792 9. 65 (Boschetti et al. , 2012) Transcriptome Caenorhabditis elegans 11168 206 1. 84 (Boschetti et al. , 2012) Transcriptome Cryptosporidium parvum 4074 8 0. 20 (Alsmark et al. , 2013) Genome Dictyostelium discoideum 13605 61, 92 0. 45, 0. 68 (Alsmark et al. , 2013; Clarke et al. , 2013) Genome Drosophila melanogaster 18368 105 0. 57 (Boschetti et al. , 2012) Transcriptome Encephalitozoon cuniculi 1918 1 0. 05 (Alsmark et al. , 2013) Genome Entamoeba histolytica 9090 51, 199 0. 56, 2. 19 (Alsmark et al. , 2013; Clarke et al. , 2013) Genome Galdieria sulphuraria 6623 75 1. 13 (Schönknecht et al. , 2013) Genome Giardia intestinalis 6394 15 0. 23 (Alsmark et al. , 2013) Genome Hydra magnipapillata ~20000 71 0. 36 (Chapman et al. , 2010) Genome Leishmania major 7111 63 0. 89 (Alsmark et al. , 2013) Genome Monosiga brevicollis 9196 139 1. 51 (Yue, Sun, Hu, & Huang, 2013) Genome Naegleria gruberi 15727 44, 431 0. 28, 2. 74 (Clarke et al. , 2013; Fritz-Laylin et al. , 2010) Genome Plasmodium falciparum 5258 18 0. 34 (Alsmark et al. , 2013) Genome Phaeodactylum tricornutum 10402 587 5. 64 (Bowler et al. , 2008) Genome Toxoplasma gondii 7793 16 0. 21 (Alsmark et al. , 2013) Genome Trichomonas vaginalis 59681 134 0. 22 (Alsmark et al. , 2013) Genome Trypanosoma brucei 9750 45 0. 46 (Alsmark et al. , 2013) Genome Trypanosoma cruzi 20184 46 0. 23 (Alsmark et al. , 2013) Genome

Directions of HGT • Prokaryota → Prokaryota (frequent, spread of antibiotic resistence led to discovery of HGT) • Prokaryota → Eukaryota (e. g. endosymbiotic gene transfer) • Eukaryota → Prokaryota (rare, limited by introns; e. g. genes for pro tubulin and aktin in bacteria) • Eukaryota → Eukaryota (less common; more frequent in fungi)

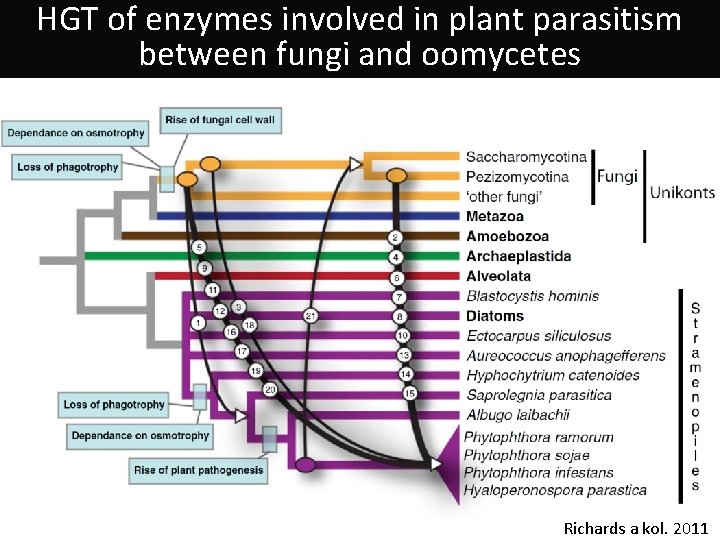
HGT of enzymes involved in plant parasitism between fungi and oomycetes Richards a kol. 2011

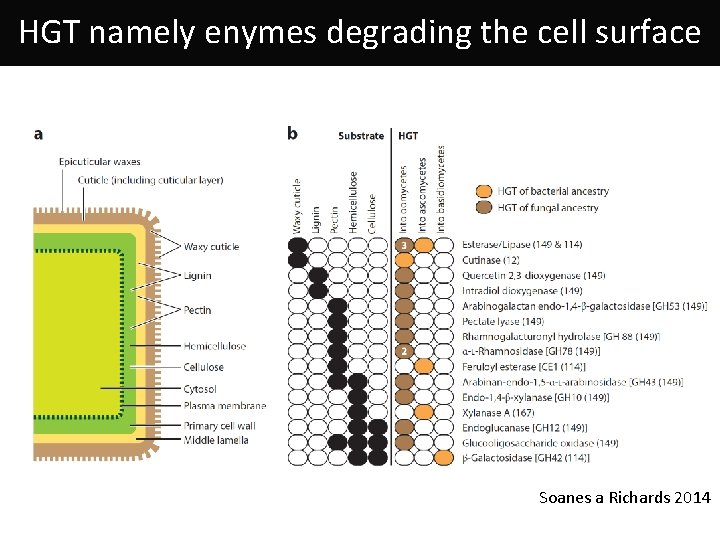
HGT namely enymes degrading the cell surface Soanes a Richards 2014

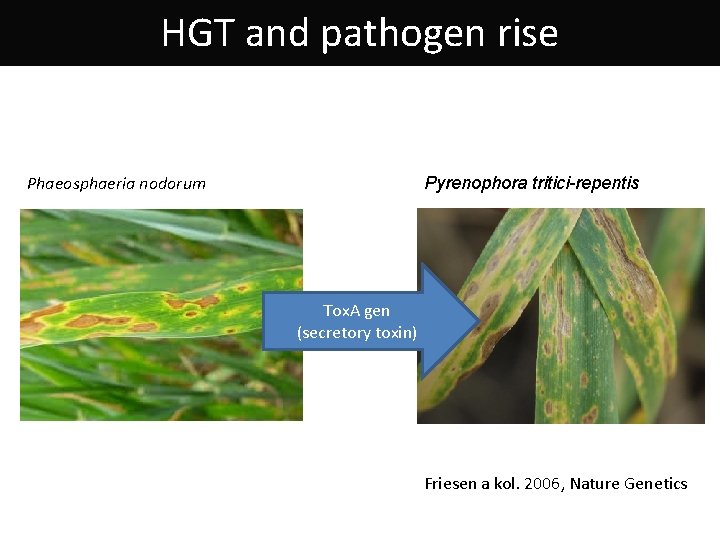
HGT and pathogen rise Phaeosphaeria nodorum Pyrenophora tritici-repentis Tox. A gen (secretory toxin) Friesen a kol. 2006, Nature Genetics

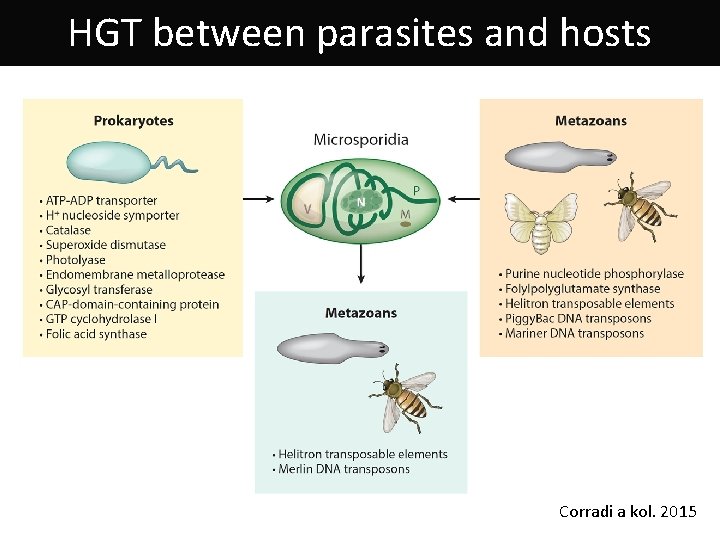
HGT between parasites and hosts Corradi a kol. 2015

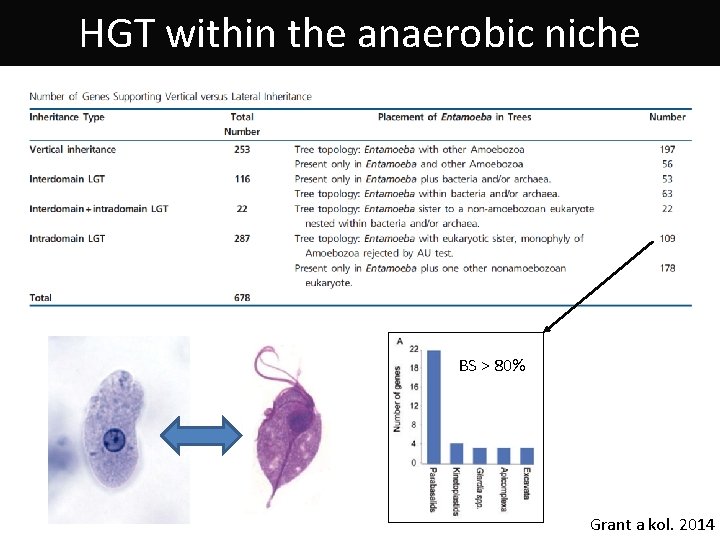
HGT within the anaerobic niche BS > 80% Grant a kol. 2014

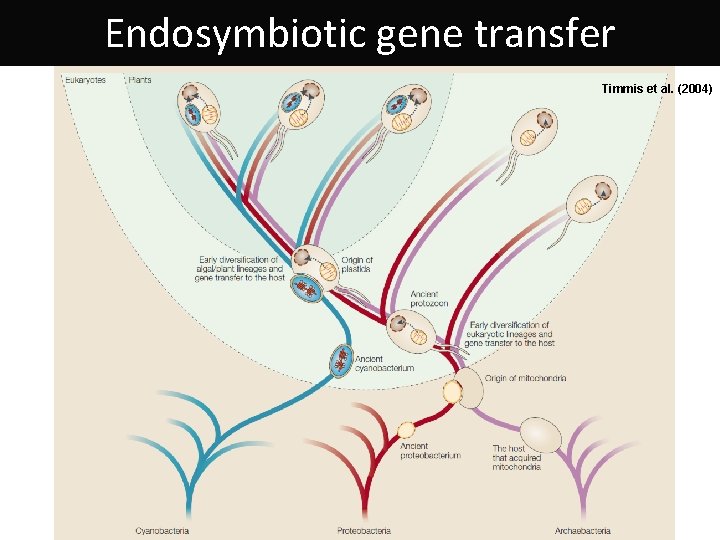
Endosymbiotic gene transfer Timmis et al. (2004)

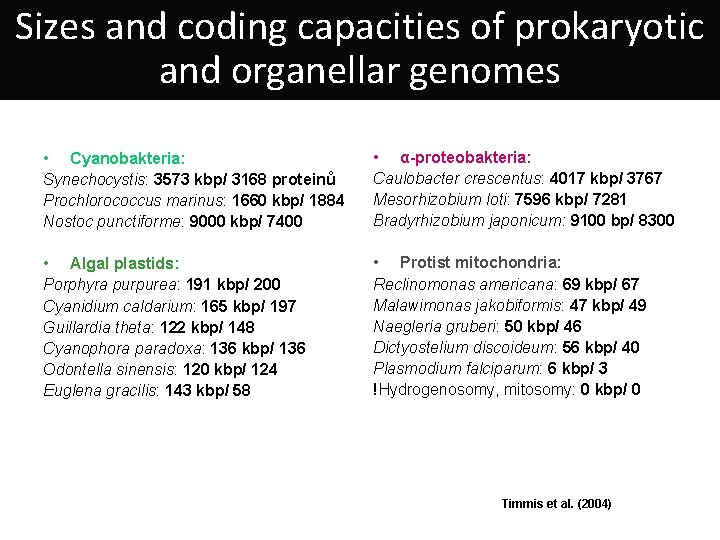
Sizes and coding capacities of prokaryotic and organellar genomes • Cyanobakteria: Synechocystis: 3573 kbp/ 3168 proteinů Prochlorococcus marinus: 1660 kbp/ 1884 Nostoc punctiforme: 9000 kbp/ 7400 • α-proteobakteria: Caulobacter crescentus: 4017 kbp/ 3767 Mesorhizobium loti: 7596 kbp/ 7281 Bradyrhizobium japonicum: 9100 bp/ 8300 • Algal plastids: Porphyra purpurea: 191 kbp/ 200 Cyanidium caldarium: 165 kbp/ 197 Guillardia theta: 122 kbp/ 148 Cyanophora paradoxa: 136 kbp/ 136 Odontella sinensis: 120 kbp/ 124 Euglena gracilis: 143 kbp/ 58 • Protist mitochondria: Reclinomonas americana: 69 kbp/ 67 Malawimonas jakobiformis: 47 kbp/ 49 Naegleria gruberi: 50 kbp/ 46 Dictyostelium discoideum: 56 kbp/ 40 Plasmodium falciparum: 6 kbp/ 3 !Hydrogenosomy, mitosomy: 0 kbp/ 0 Timmis et al. (2004)

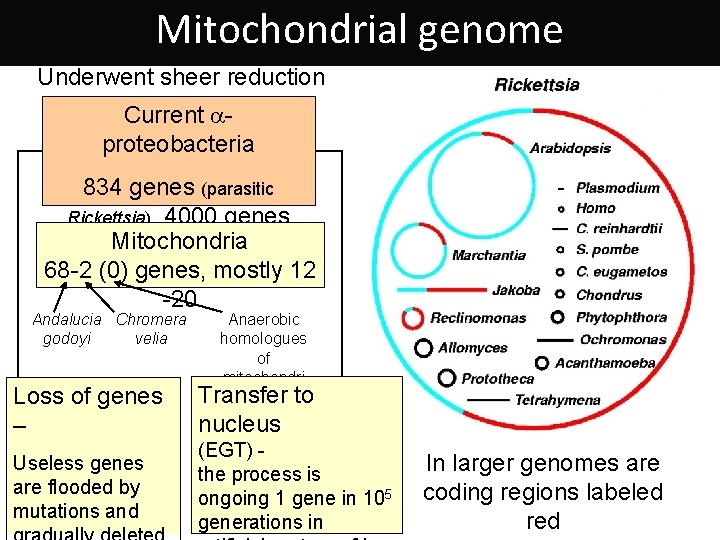
Mitochondrial genome Underwent sheer reduction Current aproteobacteria 834 genes (parasitic Rickettsia), 4000 genes (Rhodospirillum) Mitochondria 68 -2 (0) genes, mostly 12 -20 Andalucia Chromera godoyi velia Loss of genes – Useless genes are flooded by mutations and Anaerobic homologues of mitochondri a Transfer to nucleus (EGT) the process is ongoing 1 gene in 105 generations in In larger genomes are coding regions labeled red

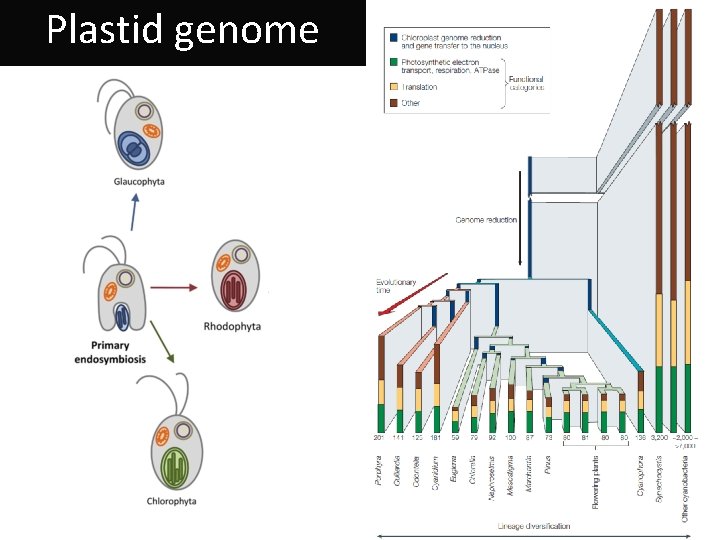
Plastid genome

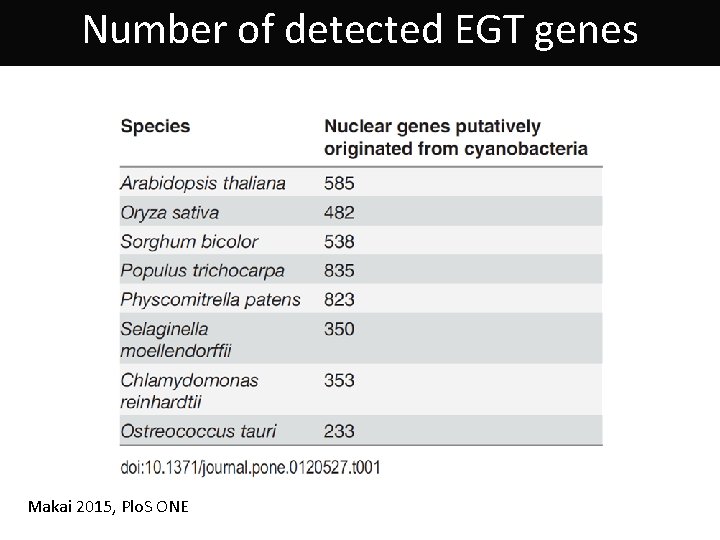
Number of detected EGT genes Makai 2015, Plo. S ONE

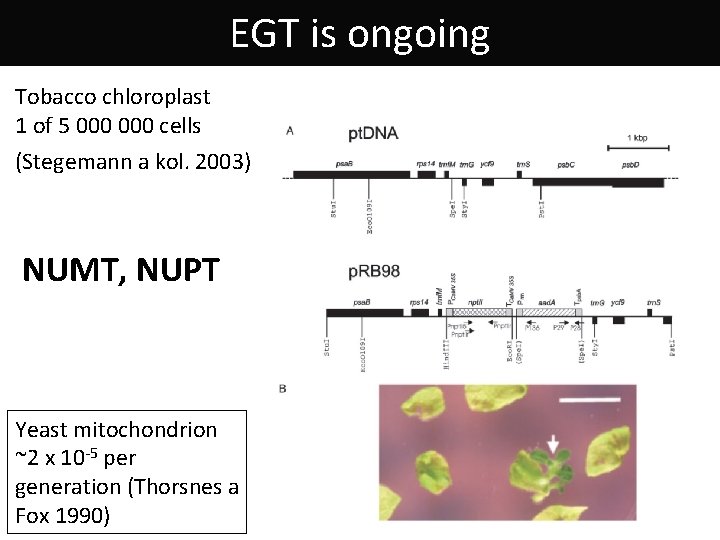
EGT is ongoing Tobacco chloroplast 1 of 5 000 cells (Stegemann a kol. 2003) NUMT, NUPT Yeast mitochondrion ~2 x 10 -5 per generation (Thorsnes a Fox 1990)

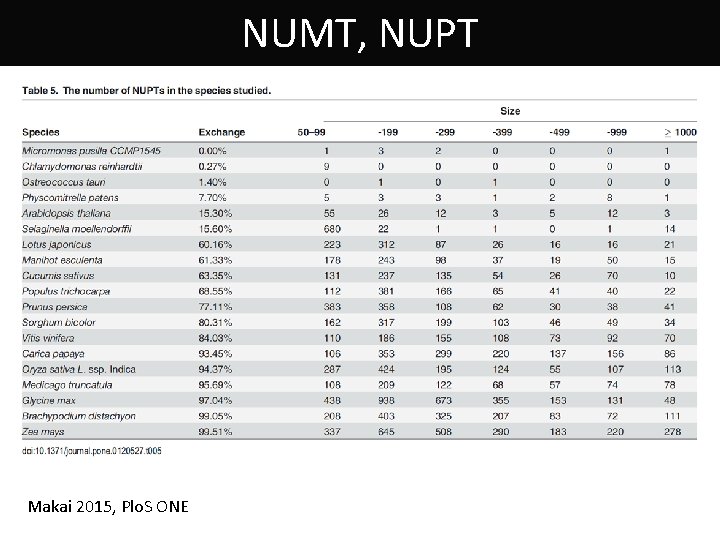
NUMT, NUPT Makai 2015, Plo. S ONE

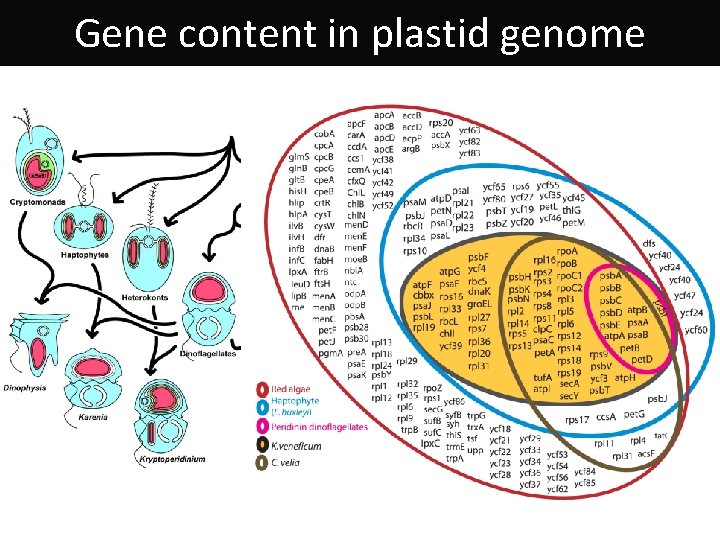
Gene content in plastid genome

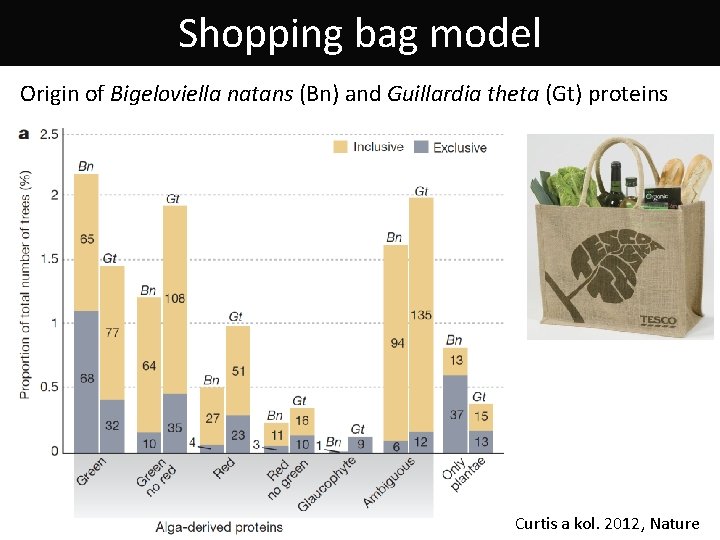
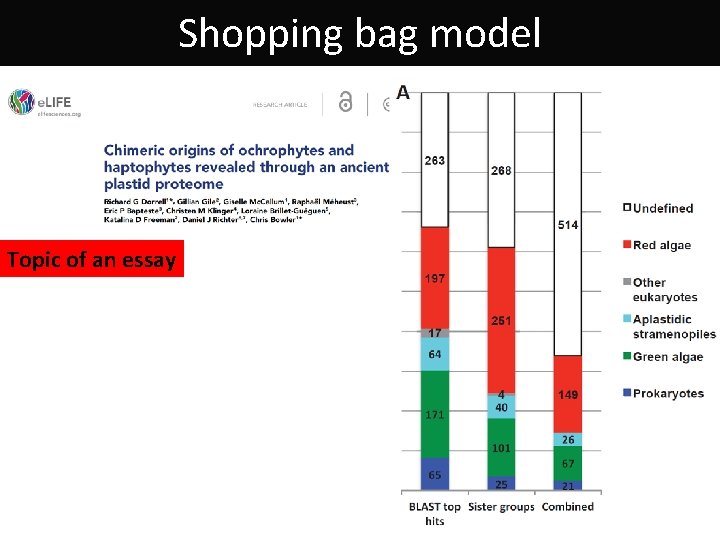
Shopping bag model Origin of Bigeloviella natans (Bn) and Guillardia theta (Gt) proteins Curtis a kol. 2012, Nature

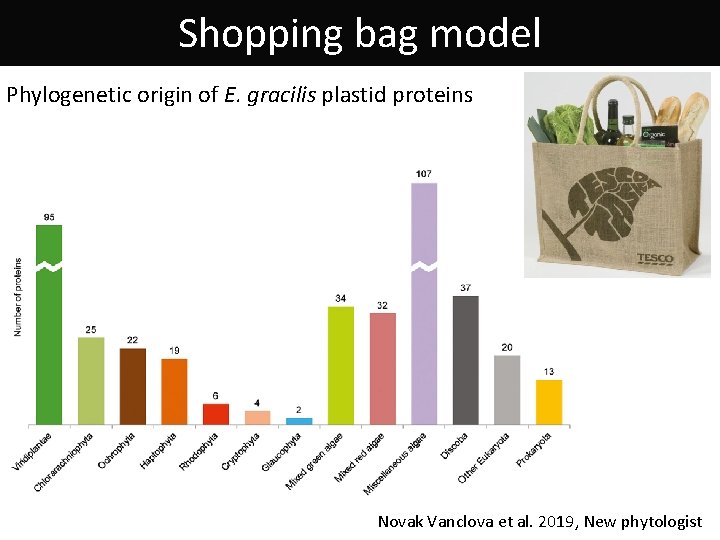
Shopping bag model Phylogenetic origin of E. gracilis plastid proteins Novak Vanclova et al. 2019, New phytologist

Shopping bag model Topic of an essay

Fraction of HGT gene in genomes • Tens of percents in bacteria • < 1% in protist genomes, not counting endosymbiotic gene transfer • ~10 -20% genes from primary endosymbiosis • < 10% genes from secondary and higher

Literature Topics of essays: Cases of HGT in protists • knihy Horizontal Gene Transfer (Genomes in Flux), 2009. • a Lateral Gene Transfer in Evolution, 2013. • Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008 Aug; 9(8): 605 -18. • Alsmark C, Foster PG, Sicheritz-Ponten T, Nakjang S, Embley TM, Hirt RP. Patterns of prokaryotic lateral gene transfers affecting parasitic microbial eukaryotes. Genome Biol. 2013 Feb 25; 14(2): R 19. • Andersson JO. Gene transfer and diversification of microbial eukaryotes. Annu Rev Microbiol. 2009; 63: 177 -93.

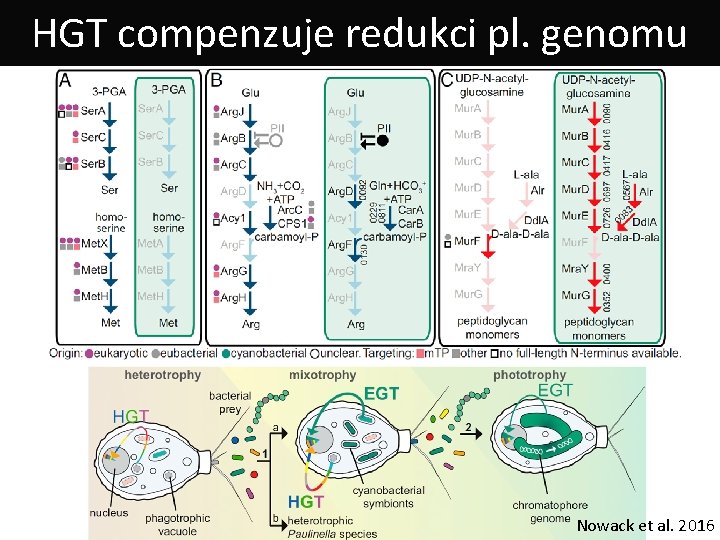
HGT compenzuje redukci pl. genomu Nowack et al. 2016

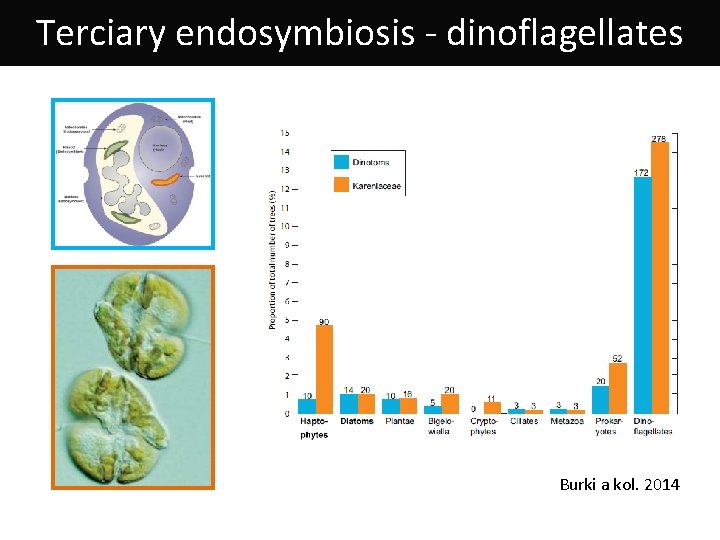
Terciary endosymbiosis - dinoflagellates Burki a kol. 2014
 Linked genes and unlinked genes
Linked genes and unlinked genes Factors of 15
Factors of 15 Linked genes and unlinked genes
Linked genes and unlinked genes Types of ferrites
Types of ferrites Weber magnetic flux
Weber magnetic flux Chapter 17: from gene to protein
Chapter 17: from gene to protein Gene by gene test results
Gene by gene test results Mechanics of heat transfer
Mechanics of heat transfer Risk transfer mechanisms
Risk transfer mechanisms Satalite weather
Satalite weather Horisontal asymptote formel
Horisontal asymptote formel Intensivering strategi
Intensivering strategi Vertikal opinionsleder
Vertikal opinionsleder Horisontal madrass sutur
Horisontal madrass sutur Horisontal
Horisontal Agrobacterium mediated gene transfer
Agrobacterium mediated gene transfer Blood transport system
Blood transport system Gene transfer by electroporation
Gene transfer by electroporation Transposase
Transposase Vertical gene transfer
Vertical gene transfer Horizontal gene transfer
Horizontal gene transfer Horizontal gene transfer
Horizontal gene transfer Vertical gene transfer
Vertical gene transfer Horizontal gene transfer
Horizontal gene transfer Horizontal gene transfer
Horizontal gene transfer Vertical gene transfer
Vertical gene transfer Horizontal gene transfer
Horizontal gene transfer Horizontal gene transfer
Horizontal gene transfer Horizontal gene transfer
Horizontal gene transfer Total surface area of a rectangular prism
Total surface area of a rectangular prism Lateral transfer of learning
Lateral transfer of learning A repeating disturbance that transfers energy
A repeating disturbance that transfers energy Genes environment
Genes environment Genes de ladybug
Genes de ladybug Caudal genetico
Caudal genetico Formulacion de la teoria cromosomica de la herencia
Formulacion de la teoria cromosomica de la herencia Genes xenologos
Genes xenologos Que son genes
Que son genes Genes schizophrenia
Genes schizophrenia Immunosuppressive drugs
Immunosuppressive drugs What is codominance
What is codominance Cdc genes
Cdc genes Genes hox
Genes hox What is the relationship between dna chromosomes and genes
What is the relationship between dna chromosomes and genes Transformation in bacteria
Transformation in bacteria Cadena molde de adn
Cadena molde de adn Anclaje
Anclaje A lac repressor turns off the lac genes by
A lac repressor turns off the lac genes by