Hierarchical Sequencing CS 273 a Lecture 4 Autumn






- Slides: 19

Hierarchical Sequencing CS 273 a Lecture 4, Autumn 08, Batzoglou

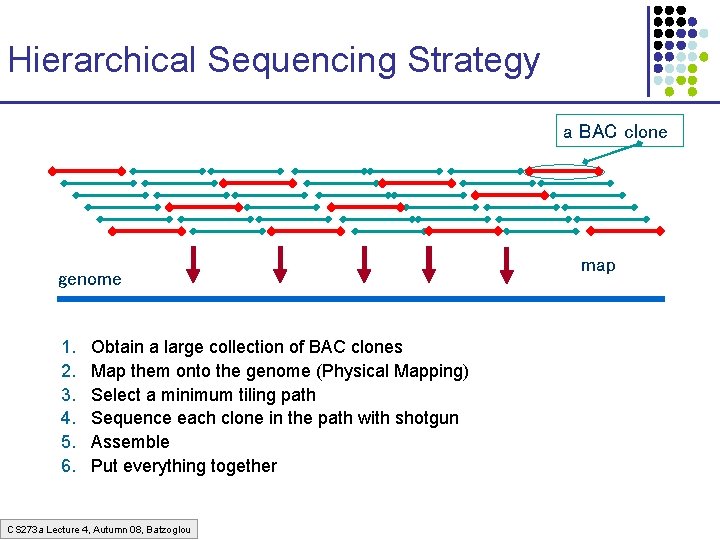
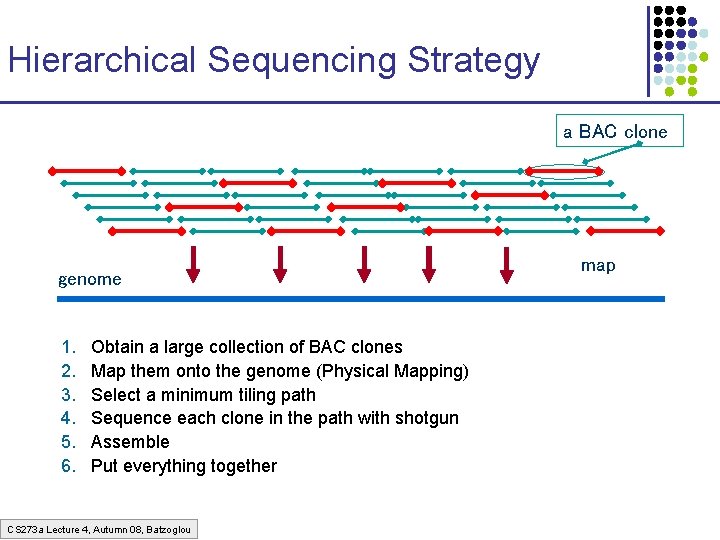
Hierarchical Sequencing Strategy a BAC clone genome 1. 2. 3. 4. 5. 6. Obtain a large collection of BAC clones Map them onto the genome (Physical Mapping) Select a minimum tiling path Sequence each clone in the path with shotgun Assemble Put everything together CS 273 a Lecture 4, Autumn 08, Batzoglou map

Hierarchical Sequencing Strategy a BAC clone genome 1. 2. 3. 4. 5. 6. Obtain a large collection of BAC clones Map them onto the genome (Physical Mapping) Select a minimum tiling path Sequence each clone in the path with shotgun Assemble Put everything together CS 273 a Lecture 4, Autumn 08, Batzoglou map

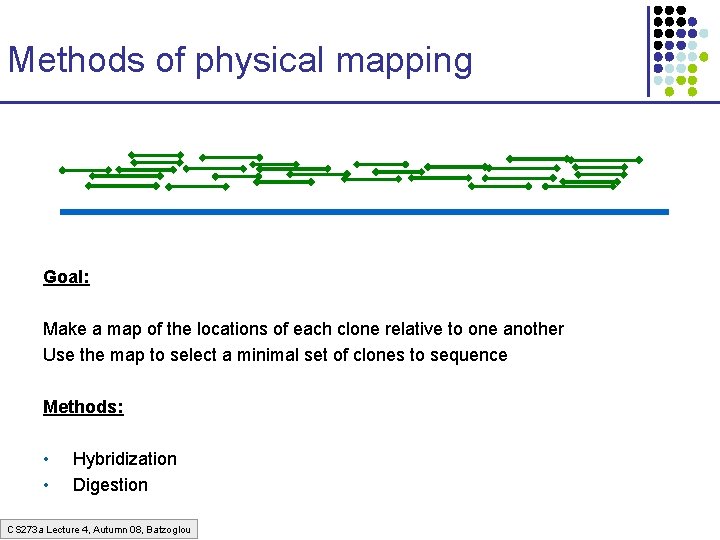
Methods of physical mapping Goal: Make a map of the locations of each clone relative to one another Use the map to select a minimal set of clones to sequence Methods: • • Hybridization Digestion CS 273 a Lecture 4, Autumn 08, Batzoglou

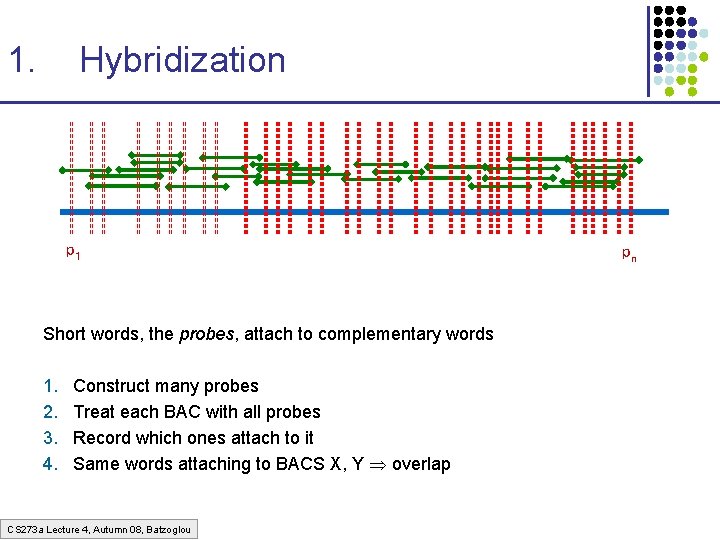
1. Hybridization p 1 Short words, the probes, attach to complementary words 1. 2. 3. 4. Construct many probes Treat each BAC with all probes Record which ones attach to it Same words attaching to BACS X, Y overlap CS 273 a Lecture 4, Autumn 08, Batzoglou pn

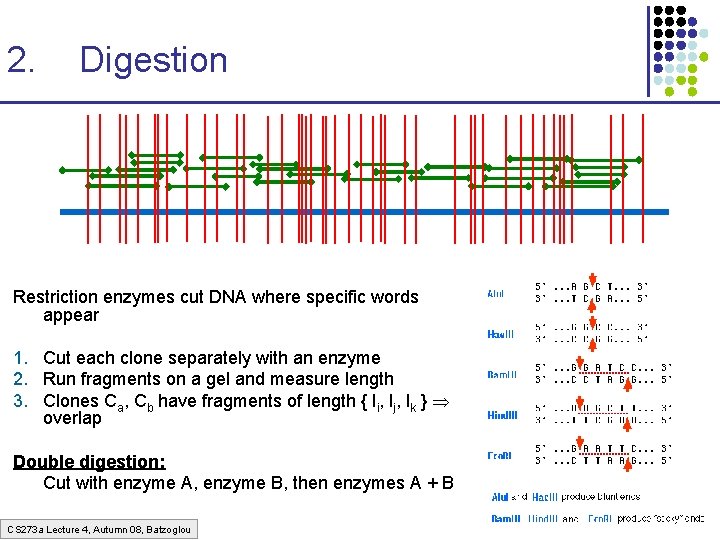
2. Digestion Restriction enzymes cut DNA where specific words appear 1. Cut each clone separately with an enzyme 2. Run fragments on a gel and measure length 3. Clones Ca, Cb have fragments of length { li, lj, lk } overlap Double digestion: Cut with enzyme A, enzyme B, then enzymes A + B CS 273 a Lecture 4, Autumn 08, Batzoglou

Online Clone-by-clone The Walking Method CS 273 a Lecture 4, Autumn 08, Batzoglou

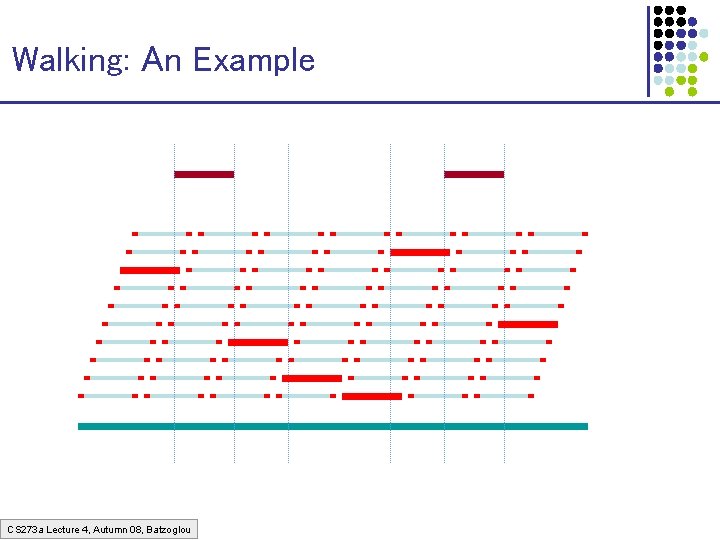
The Walking Method 1. Build a very redundant library of BACs with sequenced cloneends (cheap to build) 2. Sequence some “seed” clones 3. “Walk” from seeds using clone-ends to pick library clones that extend left & right CS 273 a Lecture 4, Autumn 08, Batzoglou

Walking: An Example CS 273 a Lecture 4, Autumn 08, Batzoglou

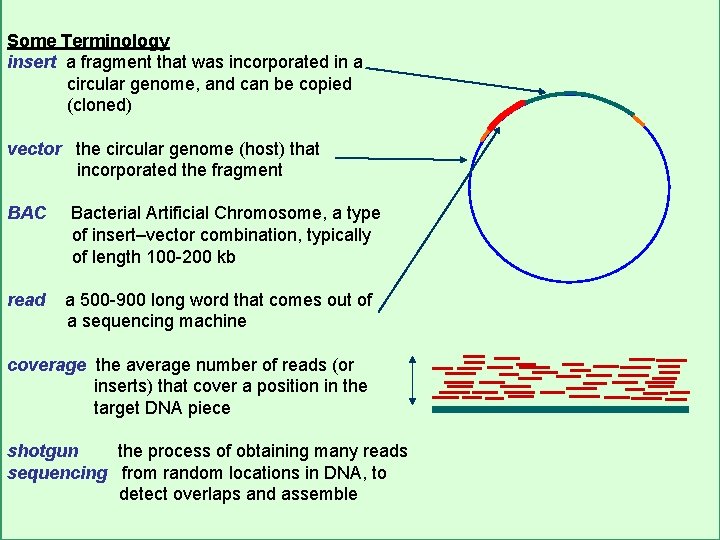
Some Terminology insert a fragment that was incorporated in a circular genome, and can be copied (cloned) vector the circular genome (host) that incorporated the fragment BAC read Bacterial Artificial Chromosome, a type of insert–vector combination, typically of length 100 -200 kb a 500 -900 long word that comes out of a sequencing machine coverage the average number of reads (or inserts) that cover a position in the target DNA piece shotgun the process of obtaining many reads sequencing from random locations in DNA, to detect overlaps and assemble CS 273 a Lecture 4, Autumn 08, Batzoglou

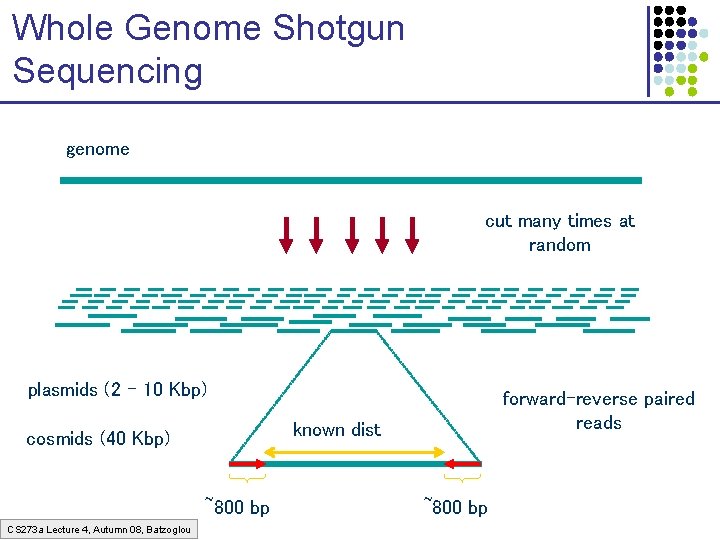
Whole Genome Shotgun Sequencing genome cut many times at random plasmids (2 – 10 Kbp) known dist cosmids (40 Kbp) ~800 bp CS 273 a Lecture 4, Autumn 08, Batzoglou forward-reverse paired reads ~800 bp

Fragment Assembly (in whole-genome shotgun sequencing) CS 273 a Lecture 4, Autumn 08, Batzoglou

Fragment Assembly Given N reads… Where N ~ 30 million… We need to use a linear-time algorithm CS 273 a Lecture 4, Autumn 08, Batzoglou

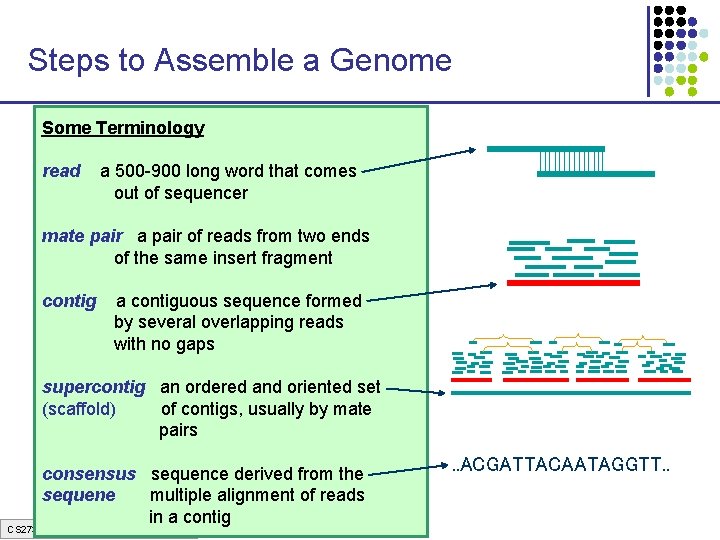
Steps to Assemble a Genome Some Terminology 1. Find overlapping readsthat comes read a 500 -900 long word out of sequencer mate pair a pair of reads from two ends 2. Merge some “good” of reads into of the same insert pairs fragment longer contigs contig a contiguous sequence formed by several overlapping reads with no gaps 3. Link contigs to form supercontigs supercontig an ordered and oriented set (scaffold) of contigs, usually by mate pairs 4. Derive consensus sequence derived from the sequene multiple alignment of reads in a contig CS 273 a Lecture 4, Autumn 08, Batzoglou . . ACGATTACAATAGGTT. .

1. Find Overlapping Reads aaactgcagtacggatct aaactgcagt … gtacggatct gggcccaaactgcagtac gggcccaaac … actgcagtac gtacggatctactacaca gtacggatct … ctactacaca CS 273 a Lecture 4, Autumn 08, Batzoglou (read, pos. , word, orient. ) (word, read, orient. , pos. ) aaactgcagt actgcagta … gtacggatct gggcccaaac gcccaaact … actgcagtac gtacggatct acggatcta … ctactacaca aaactgcagt acggatcta actgcagta cccaaactg cggatctactacac ctgcagtac gcccaaact ggcccaaac gggcccaaa gtacggatct tactacaca

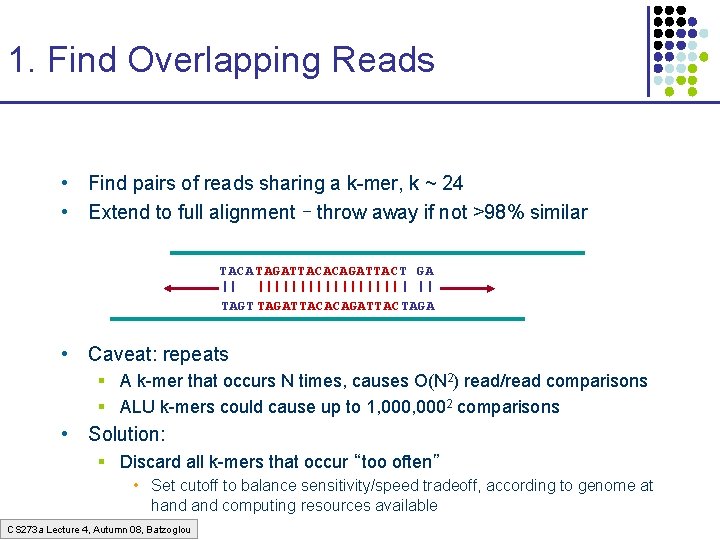
1. Find Overlapping Reads • Find pairs of reads sharing a k-mer, k ~ 24 • Extend to full alignment – throw away if not >98% similar TACA TAGATTACACAGATTAC T GA || ||||||||| | || TAGT TAGATTACACAGATTAC TAGA • Caveat: repeats § A k-mer that occurs N times, causes O(N 2) read/read comparisons § ALU k-mers could cause up to 1, 0002 comparisons • Solution: § Discard all k-mers that occur “too often” • Set cutoff to balance sensitivity/speed tradeoff, according to genome at hand computing resources available CS 273 a Lecture 4, Autumn 08, Batzoglou

1. Find Overlapping Reads Create local multiple alignments from the overlapping reads TAGATTACACAGATTACTGA TAG TTACACAGATTATTGA TAGATTACACAGATTACTGA CS 273 a Lecture 4, Autumn 08, Batzoglou

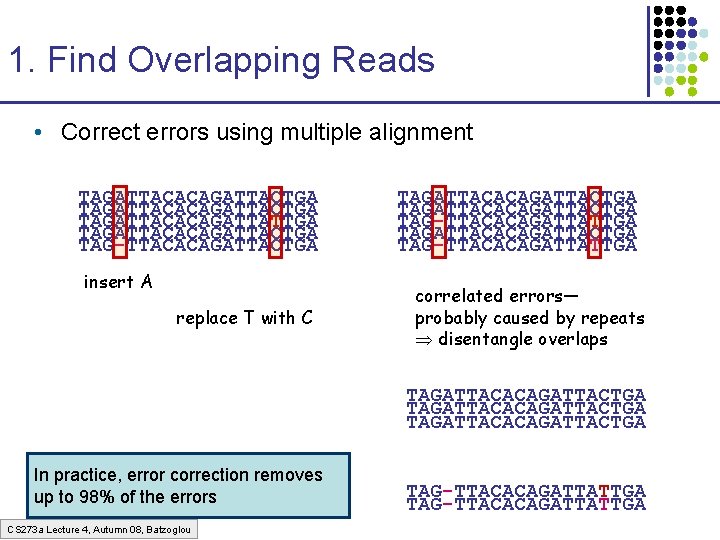
1. Find Overlapping Reads • Correct errors using multiple alignment TAGATTACACAGATTACTGA TAGATTACACAGATTATTGA TAGATTACACAGATTACTGA TAG-TTACACAGATTACTGA insert A replace T with C TAGATTACACAGATTACTGA TAG-TTACACAGATTATTGA correlated errors— probably caused by repeats disentangle overlaps TAGATTACACAGATTACTGA In practice, error correction removes up to 98% of the errors CS 273 a Lecture 4, Autumn 08, Batzoglou TAG-TTACACAGATTATTGA

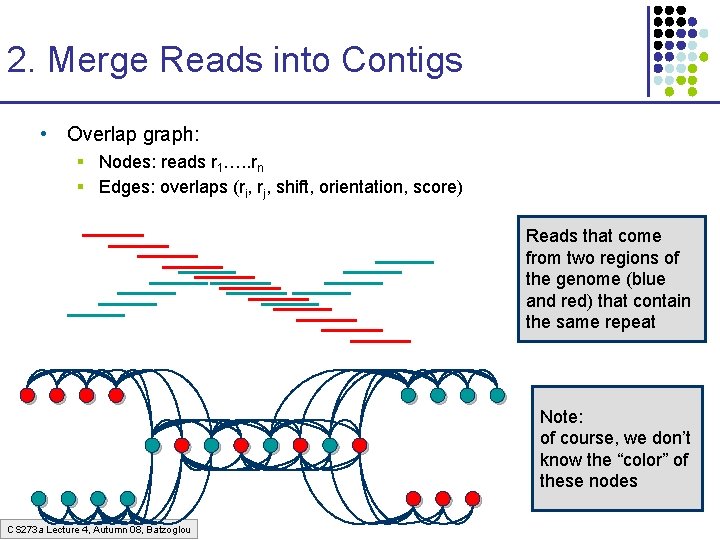
2. Merge Reads into Contigs • Overlap graph: § Nodes: reads r 1…. . rn § Edges: overlaps (ri, rj, shift, orientation, score) Reads that come from two regions of the genome (blue and red) that contain the same repeat Note: of course, we don’t know the “color” of these nodes CS 273 a Lecture 4, Autumn 08, Batzoglou