DNA Sequencing cont CS 273 a Lecture 2



- Slides: 22

DNA Sequencing (cont. ) CS 273 a Lecture 2, Autumn 10, Batzoglou

Sequencing Applications 100 million species Each individual has different DNA CS 273 a Lecture 1 CS 273 a Lecture 2, Autumn 10, Batzoglou Within individual, some cells have different DNA (i. e. cancer)

Sequencing Applications What genes are on/off, when, and in which cells? Where do molecules bind to DNA? CS 273 a Lecture 1 CS 273 a Lecture 2, Autumn 10, Batzoglou

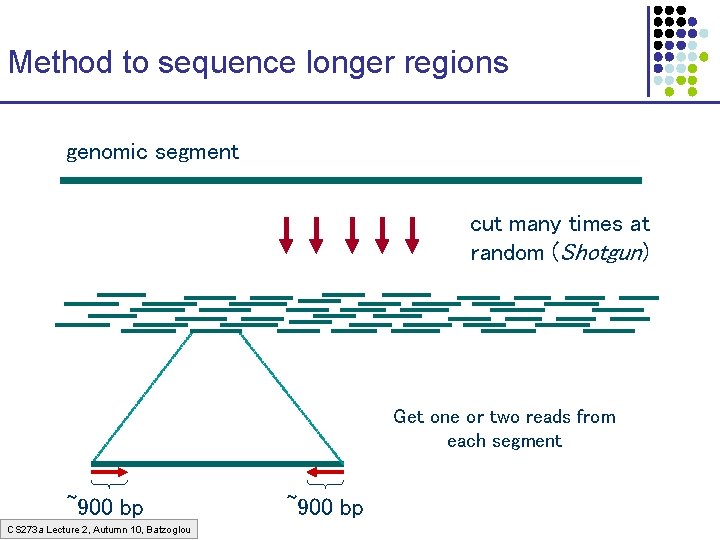
Method to sequence longer regions genomic segment cut many times at random (Shotgun) Get one or two reads from each segment ~900 bp CS 273 a Lecture 2, Autumn 10, Batzoglou ~900 bp

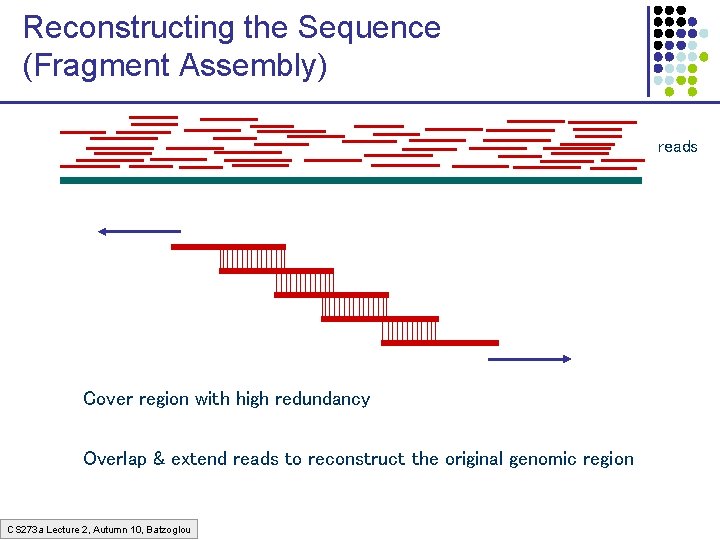
Reconstructing the Sequence (Fragment Assembly) reads Cover region with high redundancy Overlap & extend reads to reconstruct the original genomic region CS 273 a Lecture 2, Autumn 10, Batzoglou

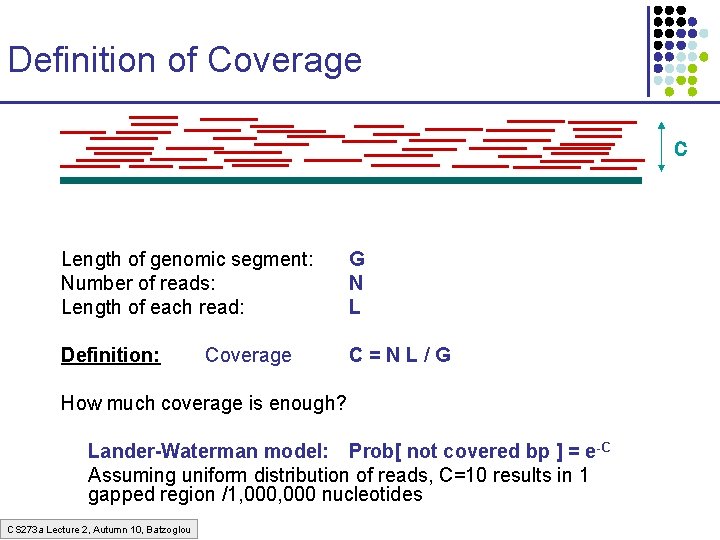
Definition of Coverage C Length of genomic segment: Number of reads: Length of each read: G N L Definition: C=NL/G Coverage How much coverage is enough? Lander-Waterman model: Prob[ not covered bp ] = e-C Assuming uniform distribution of reads, C=10 results in 1 gapped region /1, 000 nucleotides CS 273 a Lecture 2, Autumn 10, Batzoglou

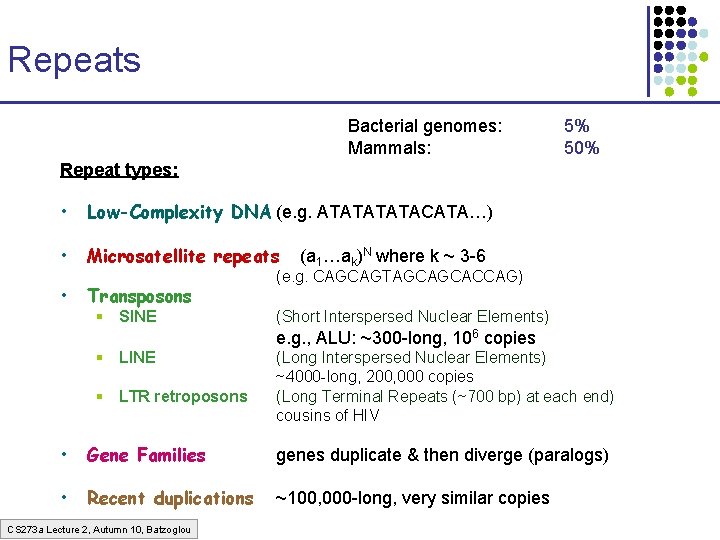
Repeats Bacterial genomes: Mammals: 5% 50% Repeat types: • Low-Complexity DNA (e. g. ATATACATA…) • Microsatellite repeats • Transposons § SINE (a 1…ak)N where k ~ 3 -6 (e. g. CAGCAGTAGCAGCACCAG) (Short Interspersed Nuclear Elements) e. g. , ALU: ~300 -long, 106 copies § LINE § LTR retroposons (Long Interspersed Nuclear Elements) ~4000 -long, 200, 000 copies (Long Terminal Repeats (~700 bp) at each end) cousins of HIV • Gene Families genes duplicate & then diverge (paralogs) • Recent duplications ~100, 000 -long, very similar copies CS 273 a Lecture 2, Autumn 10, Batzoglou

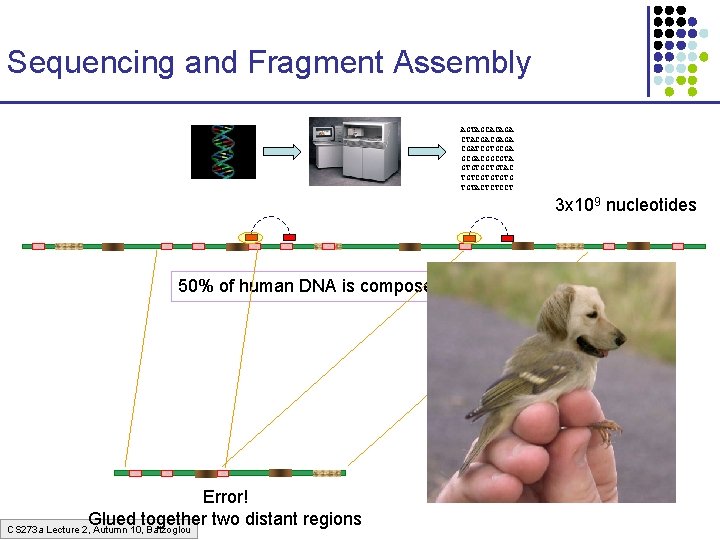
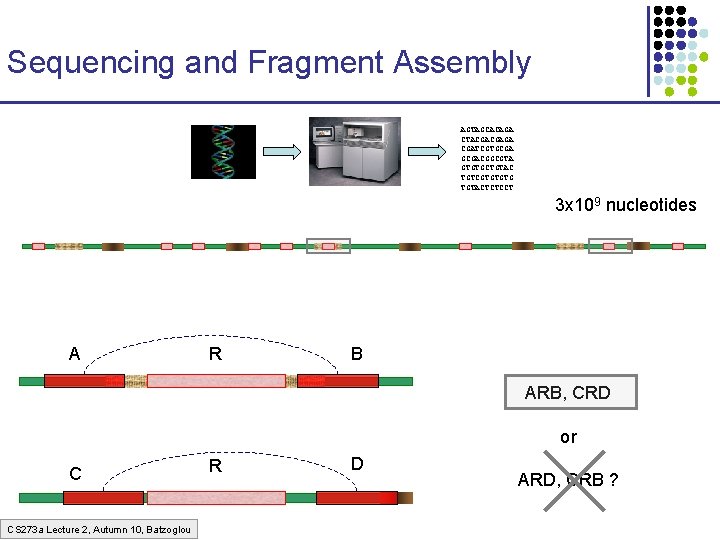
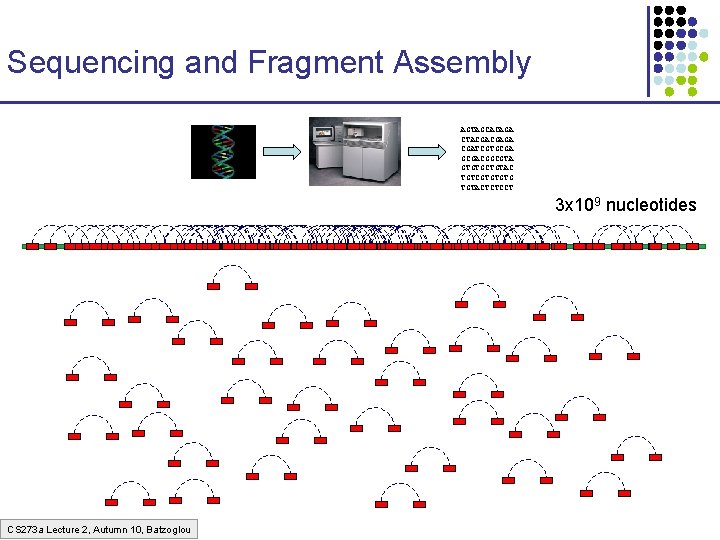
Sequencing and Fragment Assembly AGTAGCACAGA CTACGACGAGA CGATCGTGCGACGGCGTA GTGTGCTGTAC TGTCGTGTGTG TGTACTCTCCT 3 x 109 nucleotides 50% of human DNA is composed of repeats Error! Glued together two distant regions CS 273 a Lecture 2, Autumn 10, Batzoglou

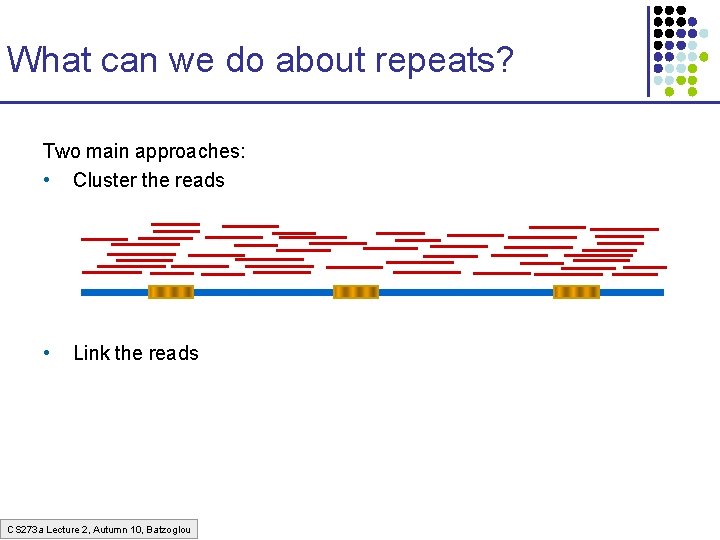
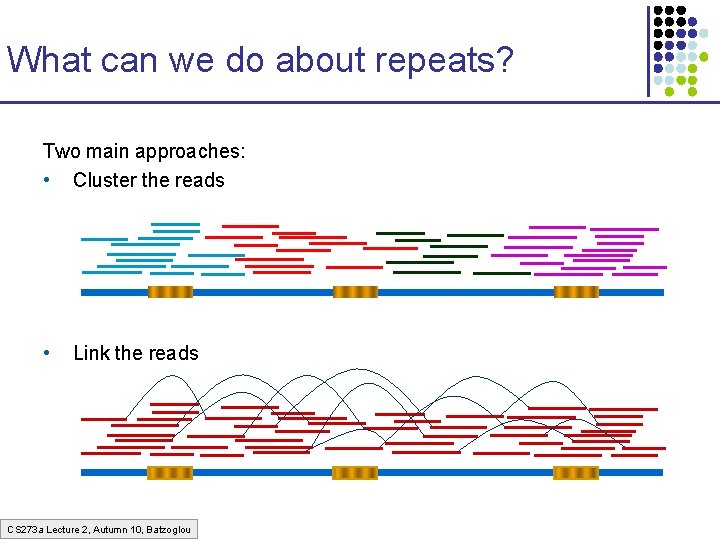
What can we do about repeats? Two main approaches: • Cluster the reads • Link the reads CS 273 a Lecture 2, Autumn 10, Batzoglou

What can we do about repeats? Two main approaches: • Cluster the reads • Link the reads CS 273 a Lecture 2, Autumn 10, Batzoglou

What can we do about repeats? Two main approaches: • Cluster the reads • Link the reads CS 273 a Lecture 2, Autumn 10, Batzoglou

Sequencing and Fragment Assembly AGTAGCACAGA CTACGACGAGA CGATCGTGCGACGGCGTA GTGTGCTGTAC TGTCGTGTGTG TGTACTCTCCT 3 x 109 nucleotides A R B ARB, CRD or C CS 273 a Lecture 2, Autumn 10, Batzoglou R D ARD, CRB ?

Sequencing and Fragment Assembly AGTAGCACAGA CTACGACGAGA CGATCGTGCGACGGCGTA GTGTGCTGTAC TGTCGTGTGTG TGTACTCTCCT 3 x 109 nucleotides CS 273 a Lecture 2, Autumn 10, Batzoglou

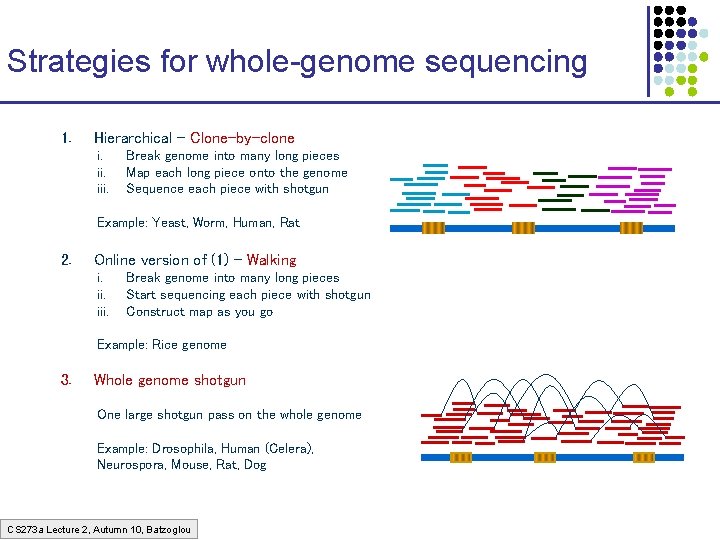
Strategies for whole-genome sequencing 1. Hierarchical – Clone-by-clone i. iii. Break genome into many long pieces Map each long piece onto the genome Sequence each piece with shotgun Example: Yeast, Worm, Human, Rat 2. Online version of (1) – Walking i. iii. Break genome into many long pieces Start sequencing each piece with shotgun Construct map as you go Example: Rice genome 3. Whole genome shotgun One large shotgun pass on the whole genome Example: Drosophila, Human (Celera), Neurospora, Mouse, Rat, Dog CS 273 a Lecture 2, Autumn 10, Batzoglou

Hierarchical Sequencing CS 273 a Lecture 2, Autumn 10, Batzoglou

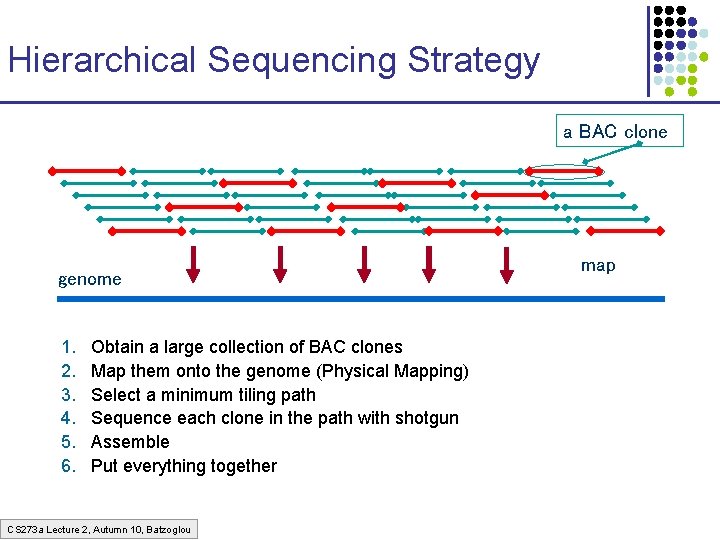
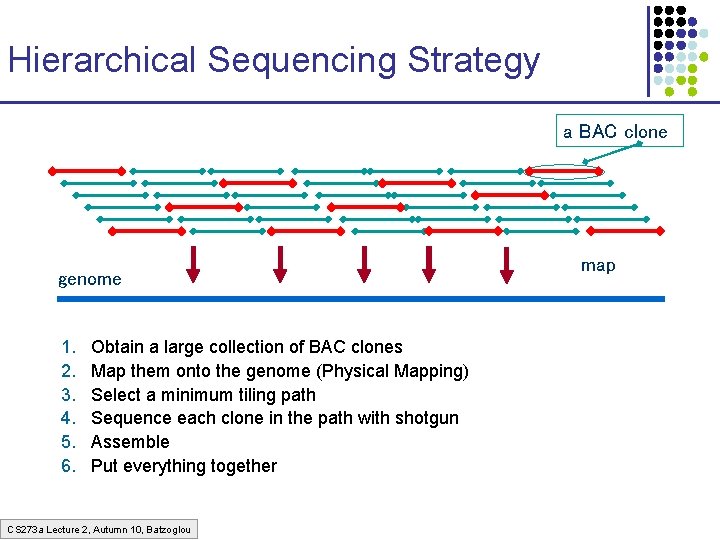
Hierarchical Sequencing Strategy a BAC clone genome 1. 2. 3. 4. 5. 6. Obtain a large collection of BAC clones Map them onto the genome (Physical Mapping) Select a minimum tiling path Sequence each clone in the path with shotgun Assemble Put everything together CS 273 a Lecture 2, Autumn 10, Batzoglou map

Hierarchical Sequencing Strategy a BAC clone genome 1. 2. 3. 4. 5. 6. Obtain a large collection of BAC clones Map them onto the genome (Physical Mapping) Select a minimum tiling path Sequence each clone in the path with shotgun Assemble Put everything together CS 273 a Lecture 2, Autumn 10, Batzoglou map

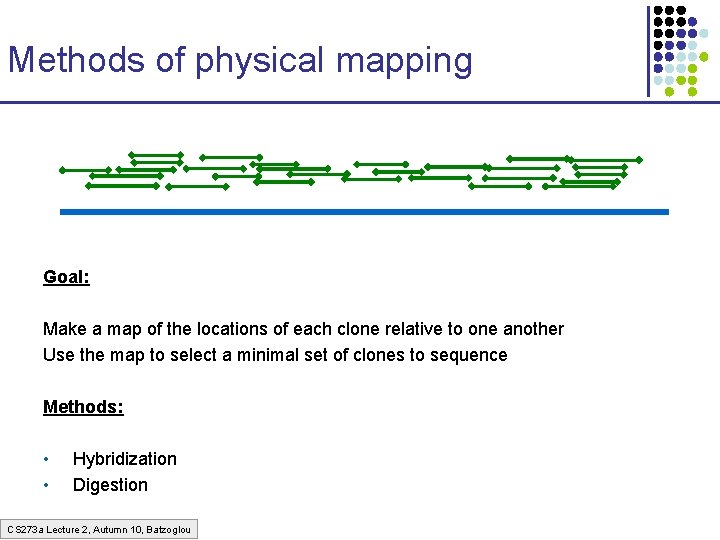
Methods of physical mapping Goal: Make a map of the locations of each clone relative to one another Use the map to select a minimal set of clones to sequence Methods: • • Hybridization Digestion CS 273 a Lecture 2, Autumn 10, Batzoglou

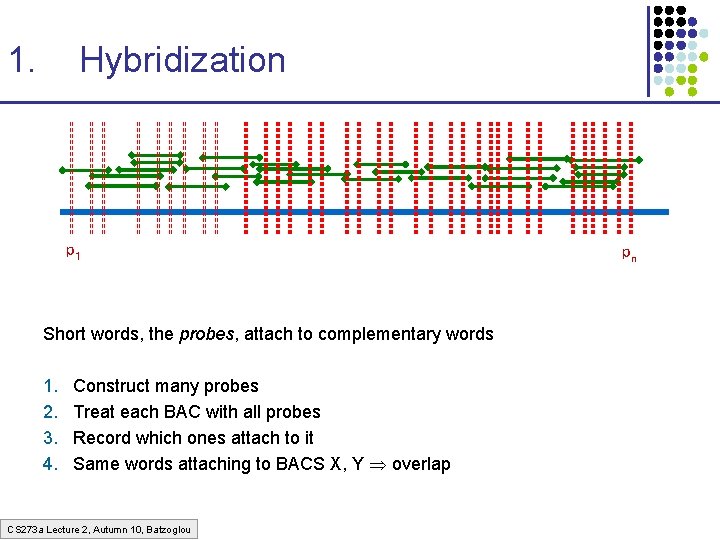
1. Hybridization p 1 Short words, the probes, attach to complementary words 1. 2. 3. 4. Construct many probes Treat each BAC with all probes Record which ones attach to it Same words attaching to BACS X, Y overlap CS 273 a Lecture 2, Autumn 10, Batzoglou pn

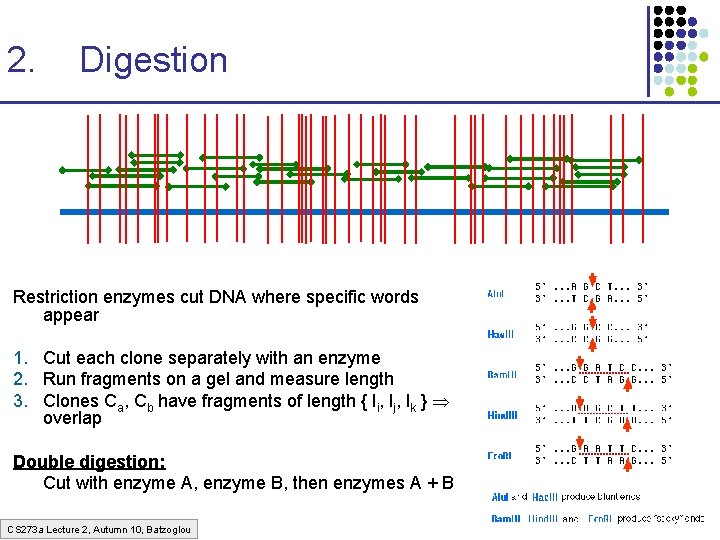
2. Digestion Restriction enzymes cut DNA where specific words appear 1. Cut each clone separately with an enzyme 2. Run fragments on a gel and measure length 3. Clones Ca, Cb have fragments of length { li, lj, lk } overlap Double digestion: Cut with enzyme A, enzyme B, then enzymes A + B CS 273 a Lecture 2, Autumn 10, Batzoglou

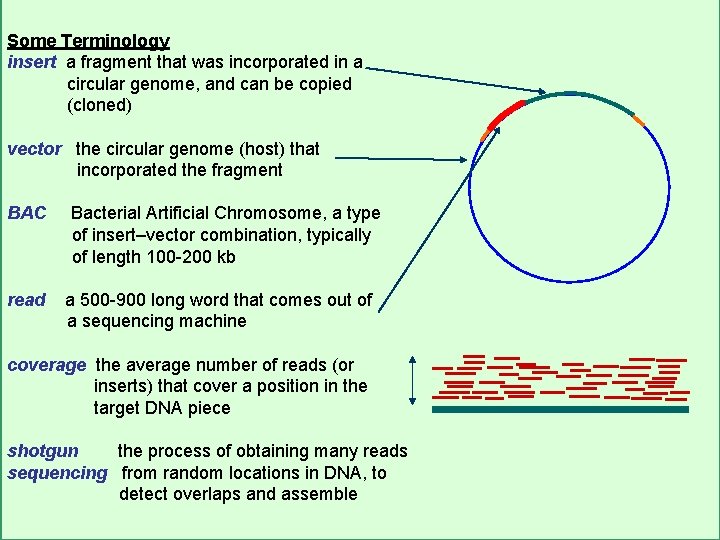
Some Terminology insert a fragment that was incorporated in a circular genome, and can be copied (cloned) vector the circular genome (host) that incorporated the fragment BAC read Bacterial Artificial Chromosome, a type of insert–vector combination, typically of length 100 -200 kb a 500 -900 long word that comes out of a sequencing machine coverage the average number of reads (or inserts) that cover a position in the target DNA piece shotgun the process of obtaining many reads sequencing from random locations in DNA, to detect overlaps and assemble CS 273 a Lecture 2, Autumn 10, Batzoglou

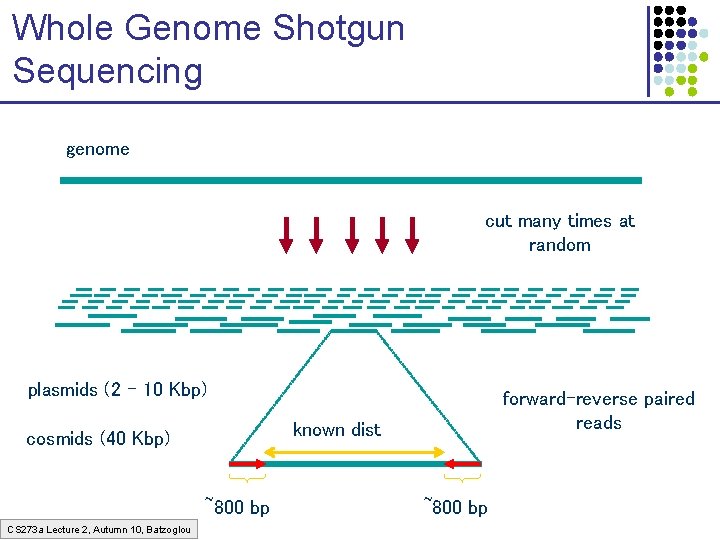
Whole Genome Shotgun Sequencing genome cut many times at random plasmids (2 – 10 Kbp) known dist cosmids (40 Kbp) ~800 bp CS 273 a Lecture 2, Autumn 10, Batzoglou forward-reverse paired reads ~800 bp