DNA Sequencing CS 273 a Lecture CS 273













- Slides: 57

DNA Sequencing CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

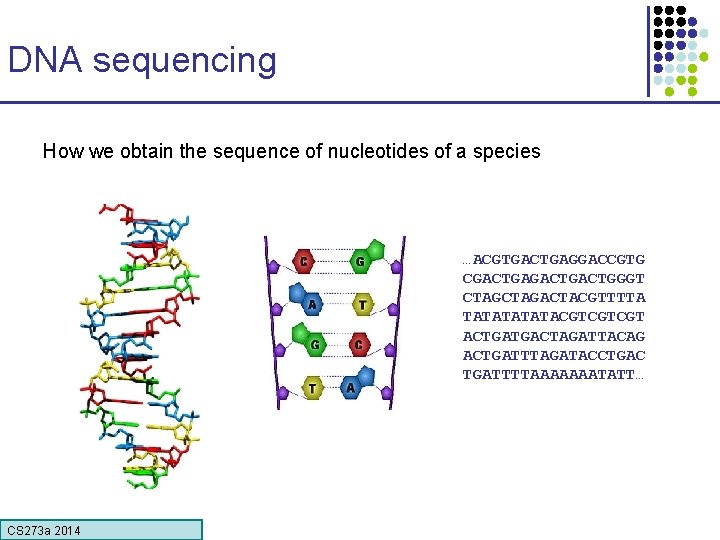
DNA sequencing How we obtain the sequence of nucleotides of a species …ACGTGACTGAGGACCGTG CGACTGACTGGGT CTAGACTACGTTTTA TATATACGTCGTCGT ACTGATGACTAGATTACAG ACTGATTTAGATACCTGAC TGATTTTAAAAAAATATT… CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

Human Genome Project 1990: Start 3 billion basepairs $3 billion 2000: Bill Clinton: 2001: Draft 2003: Finished now what? CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou “most important scientific discovery in the 20 th century”

Which representative of the species? Which human? Answer one: Answer two: it doesn’t matter Polymorphism rate: number of letter changes between two different members of a species Humans: ~1/1, 000 Other organisms have much higher polymorphism rates § Population size! CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

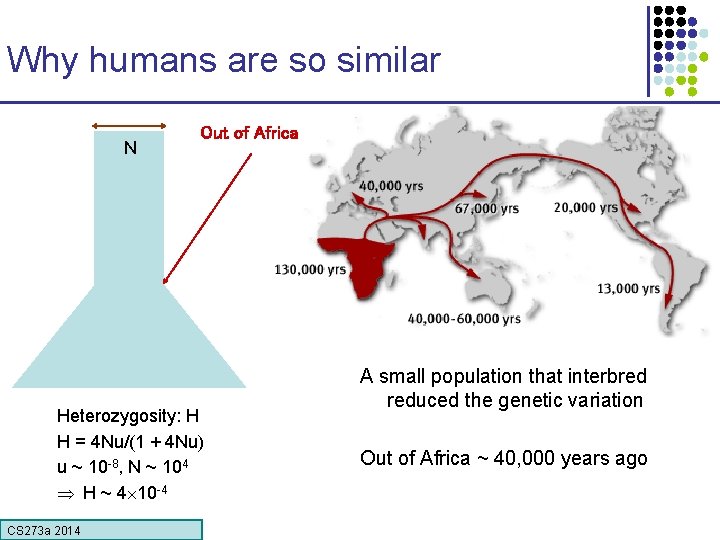
Why humans are so similar N Out of Africa Heterozygosity: H H = 4 Nu/(1 + 4 Nu) u ~ 10 -8, N ~ 104 H ~ 4 10 -4 CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou A small population that interbred reduced the genetic variation Out of Africa ~ 40, 000 years ago

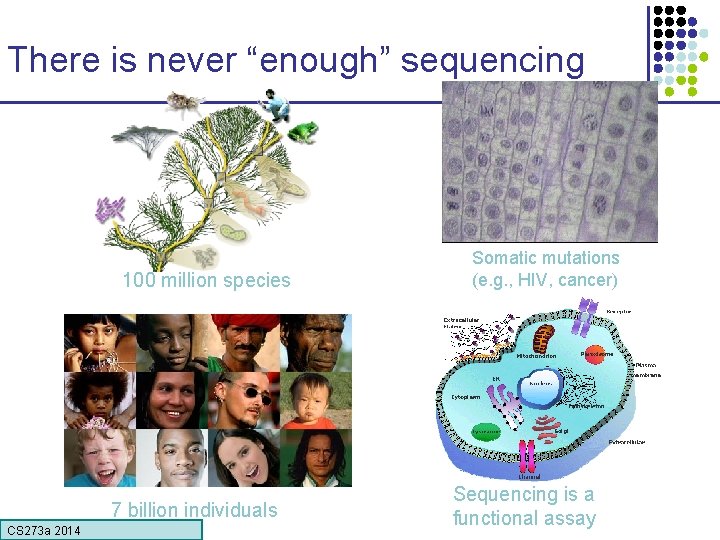
There is never “enough” sequencing 100 million species 7 billion individuals CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou Somatic mutations (e. g. , HIV, cancer) Sequencing is a functional assay

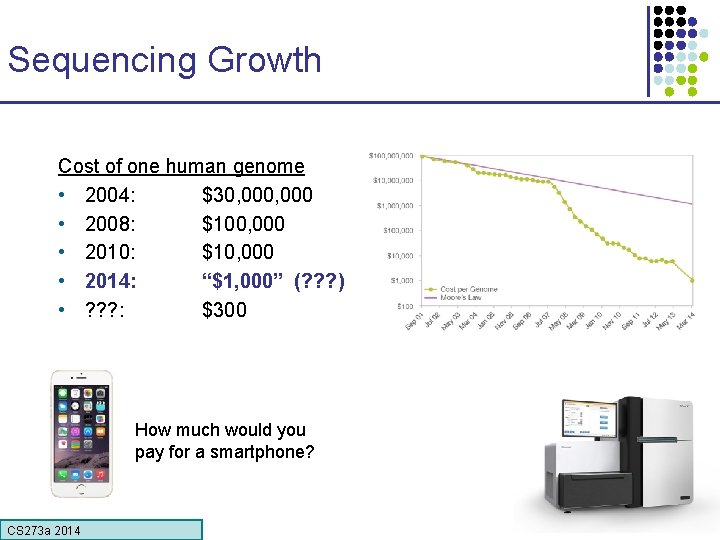
Sequencing Growth Cost of one human genome • 2004: $30, 000 • 2008: $100, 000 • 2010: $10, 000 • 2014: “$1, 000” (? ? ? ) • ? ? ? : $300 How much would you pay for a smartphone? CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

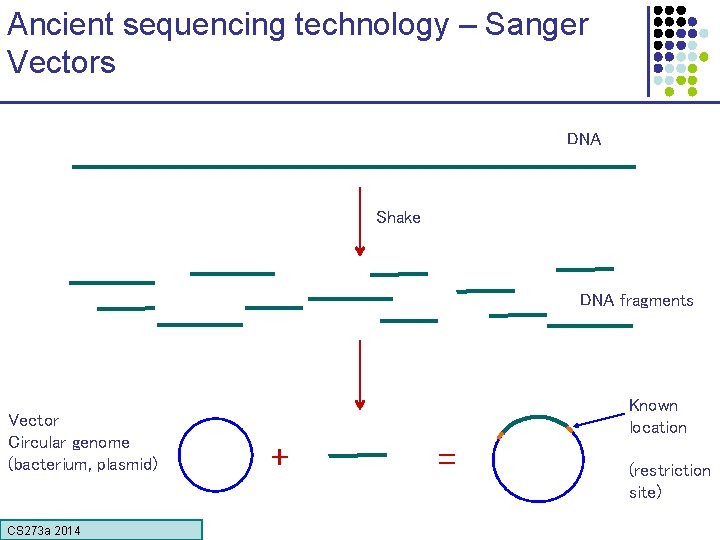
Ancient sequencing technology – Sanger Vectors DNA Shake DNA fragments Vector Circular genome (bacterium, plasmid) CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou Known location + = (restriction site)

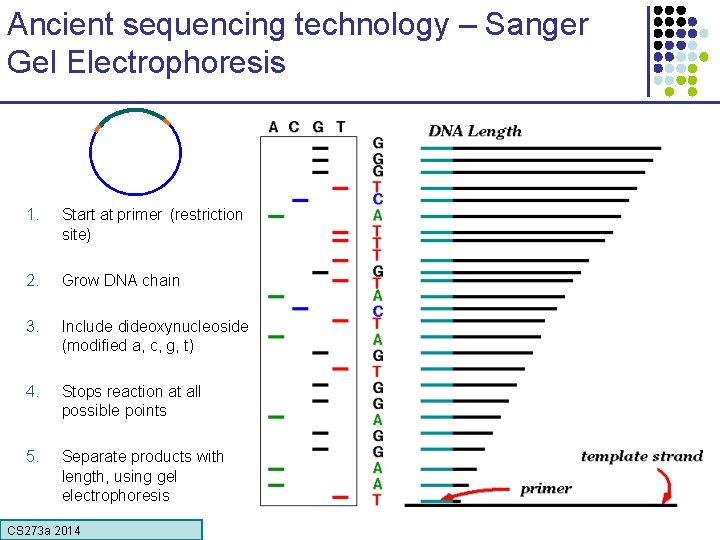
Ancient sequencing technology – Sanger Gel Electrophoresis 1. Start at primer (restriction site) 2. Grow DNA chain 3. Include dideoxynucleoside (modified a, c, g, t) 4. Stops reaction at all possible points 5. Separate products with length, using gel electrophoresis CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

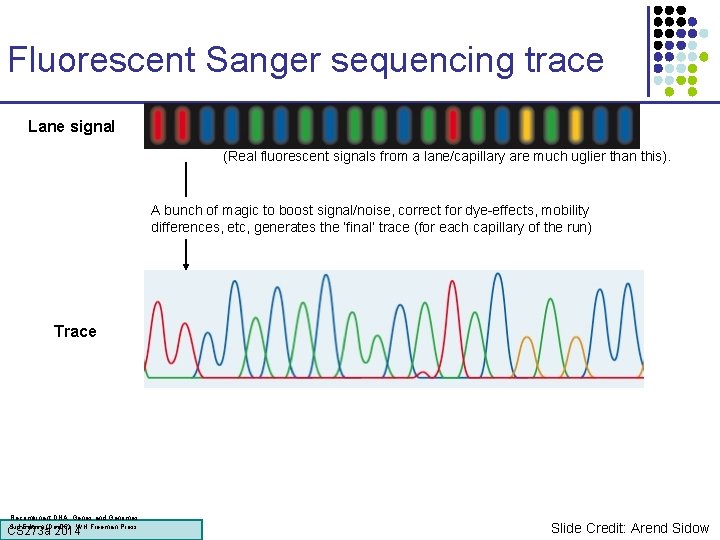
Fluorescent Sanger sequencing trace Lane signal (Real fluorescent signals from a lane/capillary are much uglier than this). A bunch of magic to boost signal/noise, correct for dye-effects, mobility differences, etc, generates the ‘final’ trace (for each capillary of the run) Trace Recombinant DNA: Genes and Genomes. 3 rd Edition (Dec 06). WH Freeman Press. CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou Slide Credit: Arend Sidow

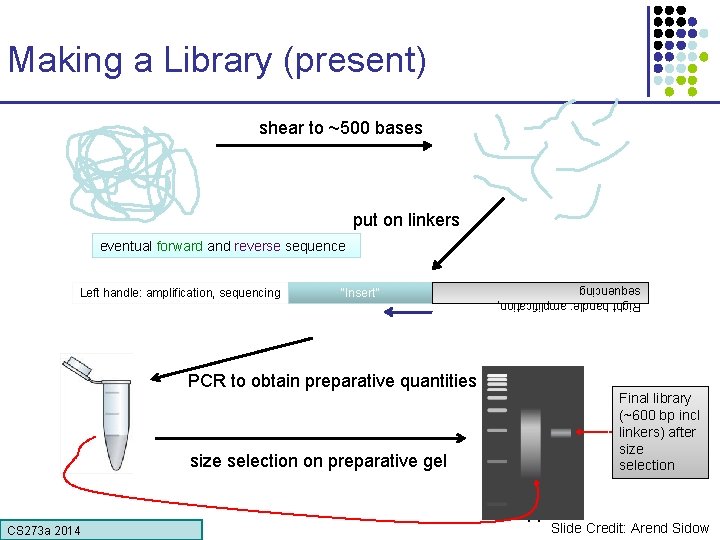
Making a Library (present) shear to ~500 bases put on linkers eventual forward and reverse sequence “Insert” Right handle: amplification, sequencing Left handle: amplification, sequencing PCR to obtain preparative quantities Final library (~600 bp incl linkers) after size selection on preparative gel CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou 11 Slide Credit: Arend Sidow

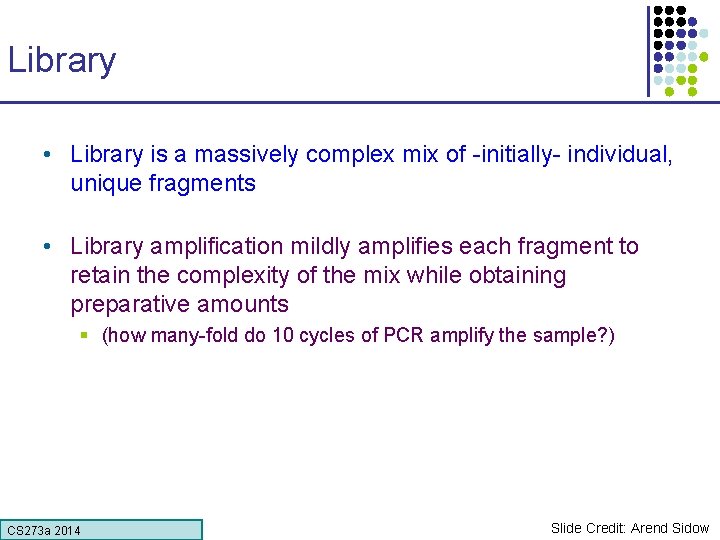
Library • Library is a massively complex mix of -initially- individual, unique fragments • Library amplification mildly amplifies each fragment to retain the complexity of the mix while obtaining preparative amounts § (how many-fold do 10 cycles of PCR amplify the sample? ) CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou Slide Credit: Arend Sidow

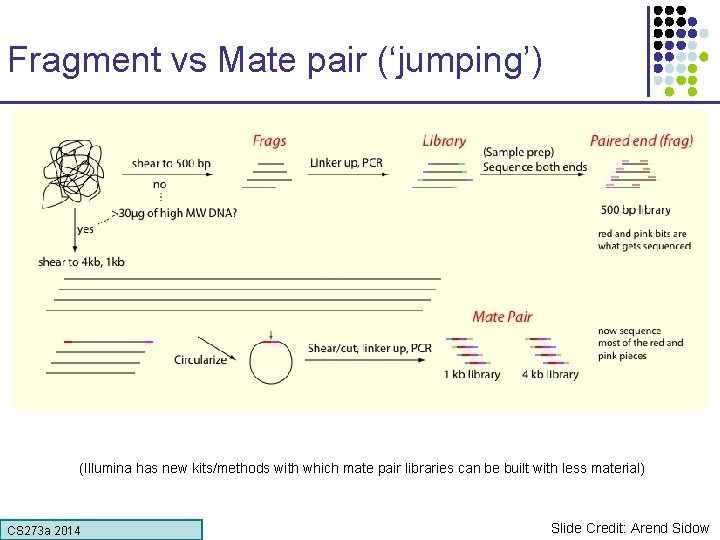
Fragment vs Mate pair (‘jumping’) (Illumina has new kits/methods with which mate pair libraries can be built with less material) CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou Slide Credit: Arend Sidow

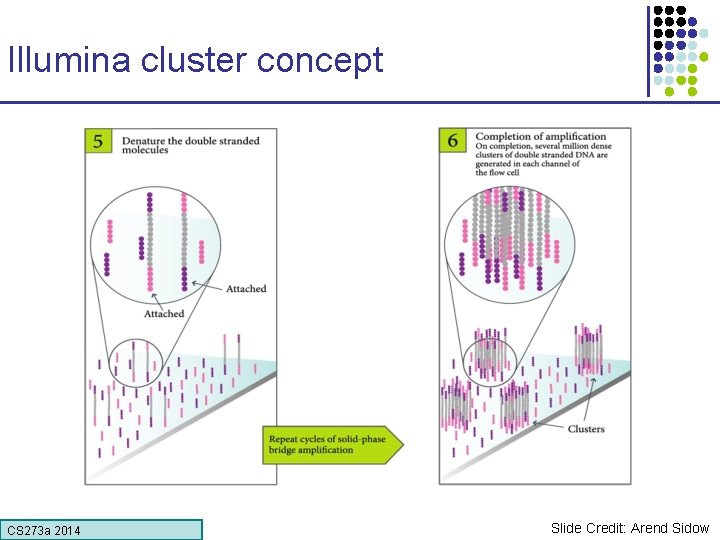
Illumina cluster concept CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou Slide Credit: Arend Sidow

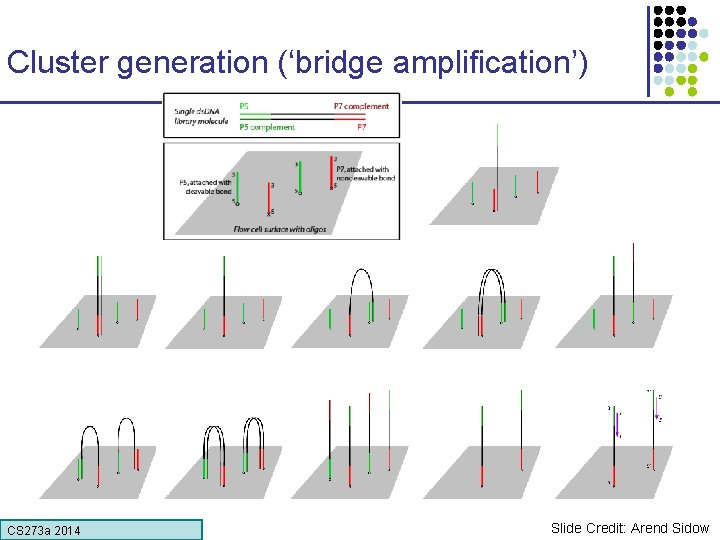
Cluster generation (‘bridge amplification’) CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou Slide Credit: Arend Sidow

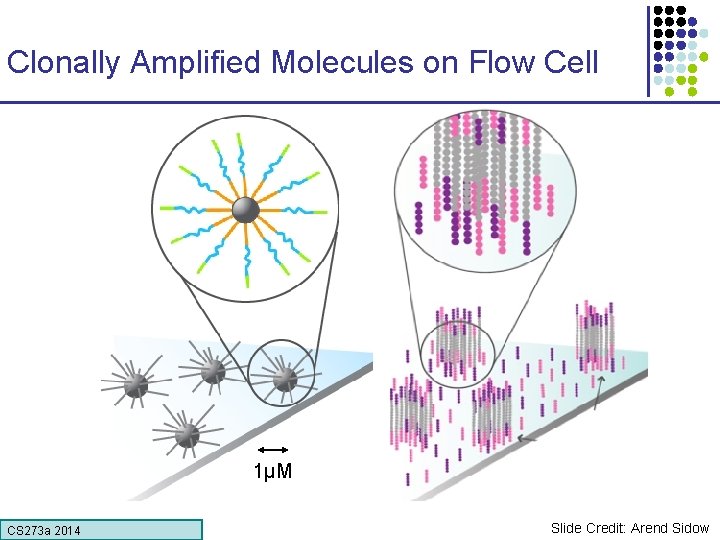
Clonally Amplified Molecules on Flow Cell 1µM CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou Slide Credit: Arend Sidow

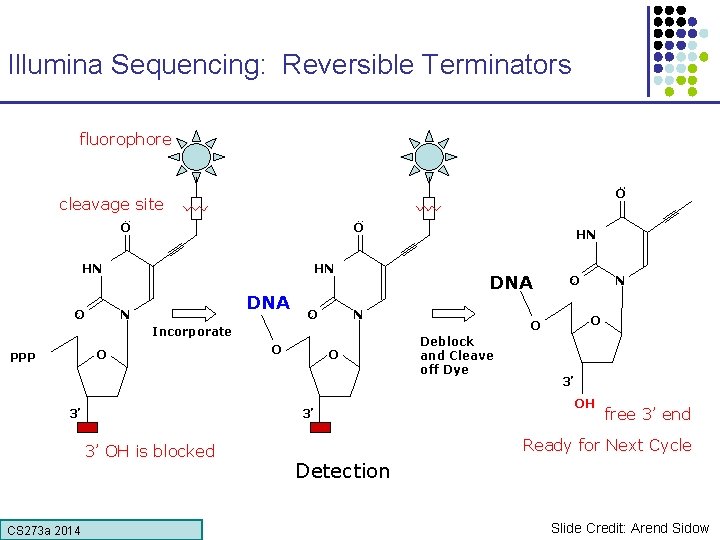
Illumina Sequencing: Reversible Terminators fluorophore O cleavage site O O HN HN O DNA N Incorporate O PPP 3’ O O 3’ 3’ OH is blocked CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou HN O O O Deblock and Cleave off Dye N 3’ OH free 3’ end Ready for Next Cycle Detection Slide Credit: Arend Sidow

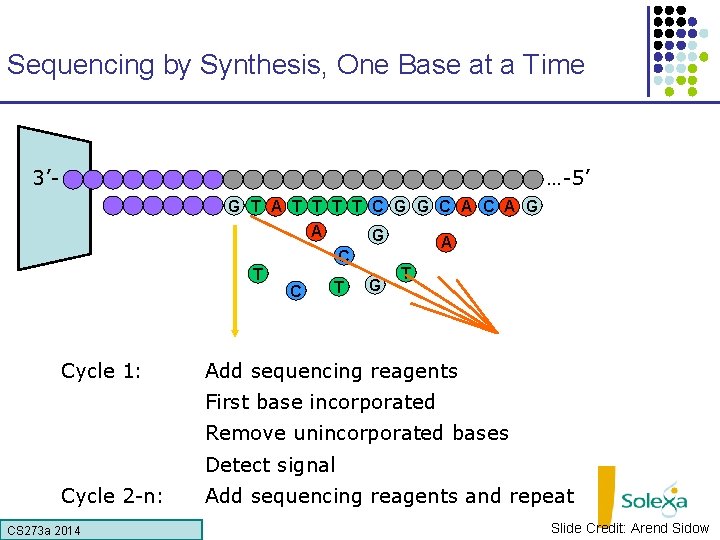
Sequencing by Synthesis, One Base at a Time 3’- …-5’ G T A T T C G G C A G A G C T Cycle 1: C T G A T Add sequencing reagents First base incorporated Remove unincorporated bases Detect signal Cycle 2 -n: CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou Add sequencing reagents and repeat Slide Credit: Arend Sidow

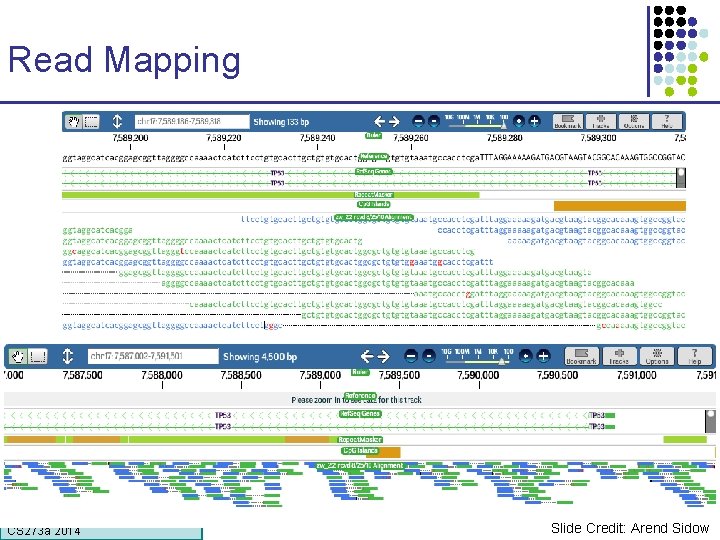
Read Mapping CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou Slide Credit: Arend Sidow

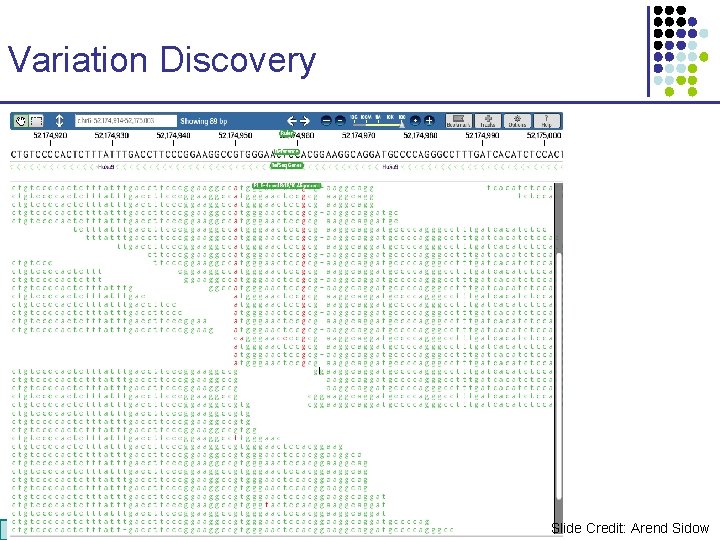
Variation Discovery CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou Slide Credit: Arend Sidow

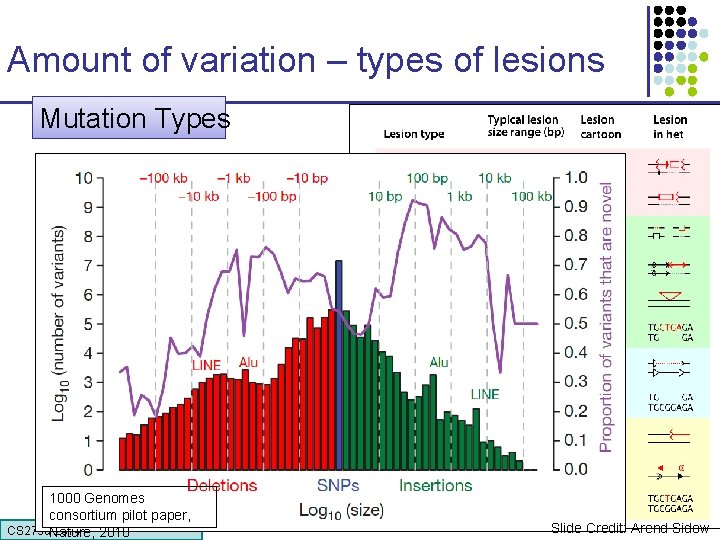
Amount of variation – types of lesions Mutation Types “we’re heterozygous in every thousandth base of our genome” 1000 Genomes 3, 000 consortium pilot paper, CS 273 a Lecture CS 273 a 2014 4, Autumn Nature, 2010 08, Batzoglou 21 Slide Credit: Arend Sidow

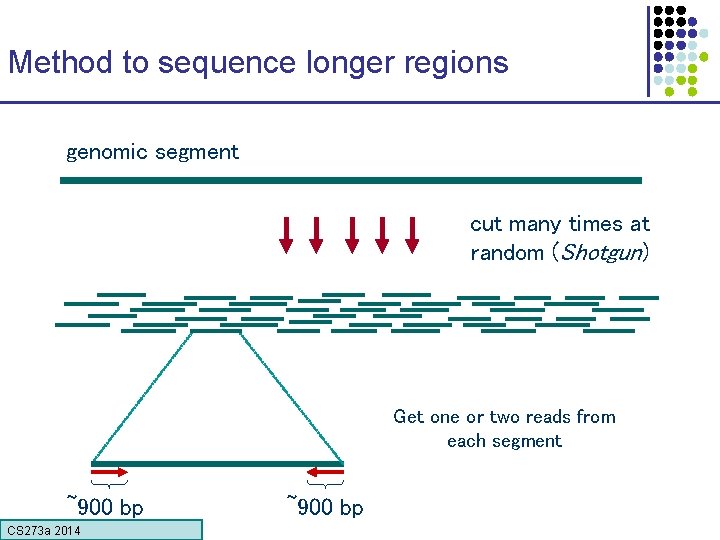
Method to sequence longer regions genomic segment cut many times at random (Shotgun) Get one or two reads from each segment ~900 bp CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou ~900 bp

Two main assembly problems • De Novo Assembly • Resequencing CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

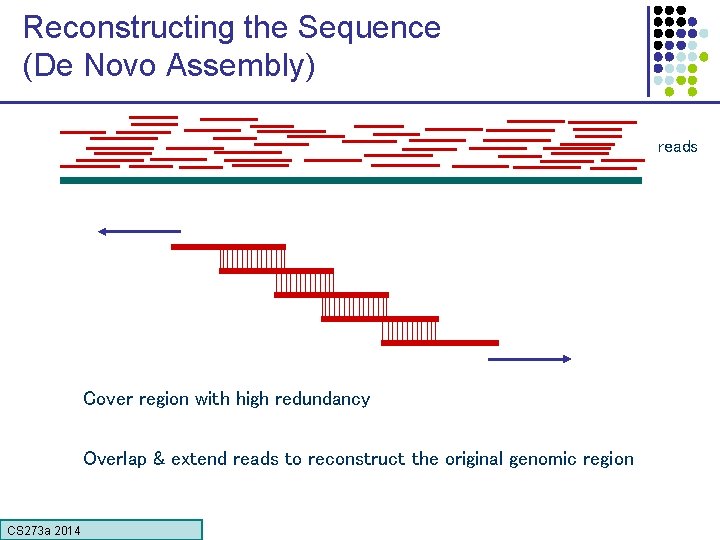
Reconstructing the Sequence (De Novo Assembly) reads Cover region with high redundancy Overlap & extend reads to reconstruct the original genomic region CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

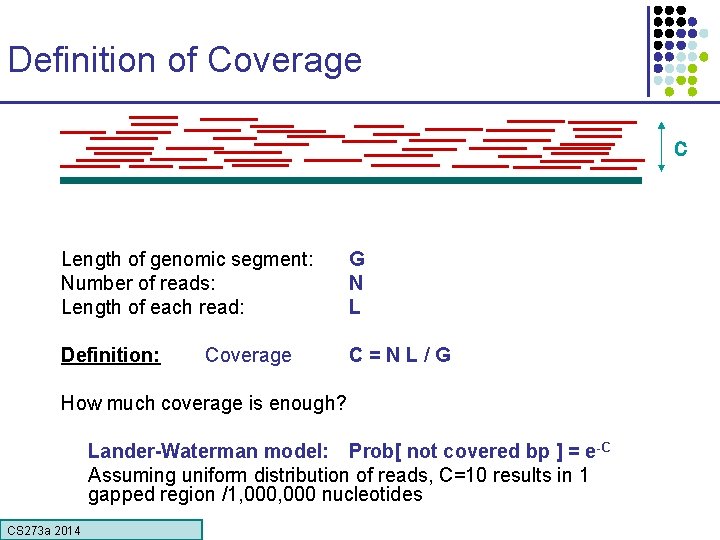
Definition of Coverage C Length of genomic segment: Number of reads: Length of each read: G N L Definition: C=NL/G Coverage How much coverage is enough? Lander-Waterman model: Prob[ not covered bp ] = e-C Assuming uniform distribution of reads, C=10 results in 1 gapped region /1, 000 nucleotides CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

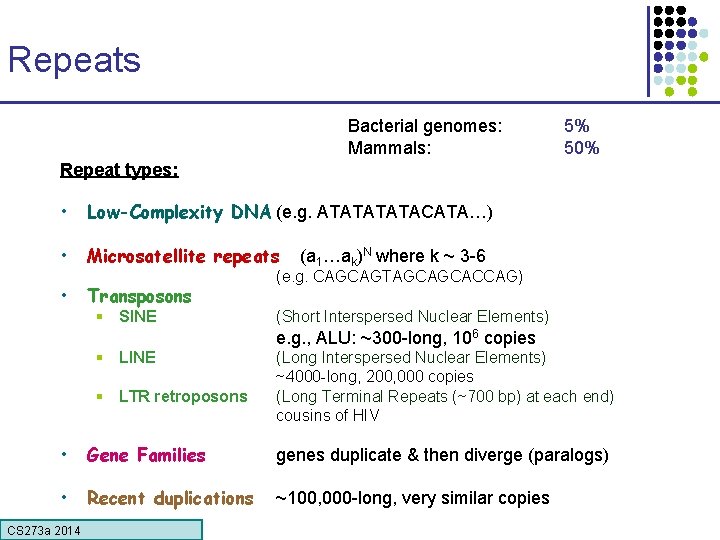
Repeats Bacterial genomes: Mammals: 5% 50% Repeat types: • Low-Complexity DNA (e. g. ATATACATA…) • Microsatellite repeats • Transposons § SINE (a 1…ak)N where k ~ 3 -6 (e. g. CAGCAGTAGCAGCACCAG) (Short Interspersed Nuclear Elements) e. g. , ALU: ~300 -long, 106 copies § LINE § LTR retroposons (Long Interspersed Nuclear Elements) ~4000 -long, 200, 000 copies (Long Terminal Repeats (~700 bp) at each end) cousins of HIV • Gene Families genes duplicate & then diverge (paralogs) • Recent duplications ~100, 000 -long, very similar copies CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

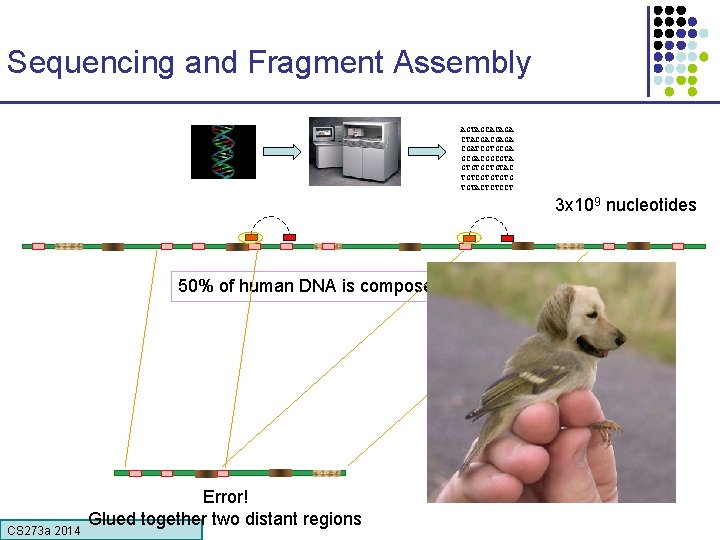
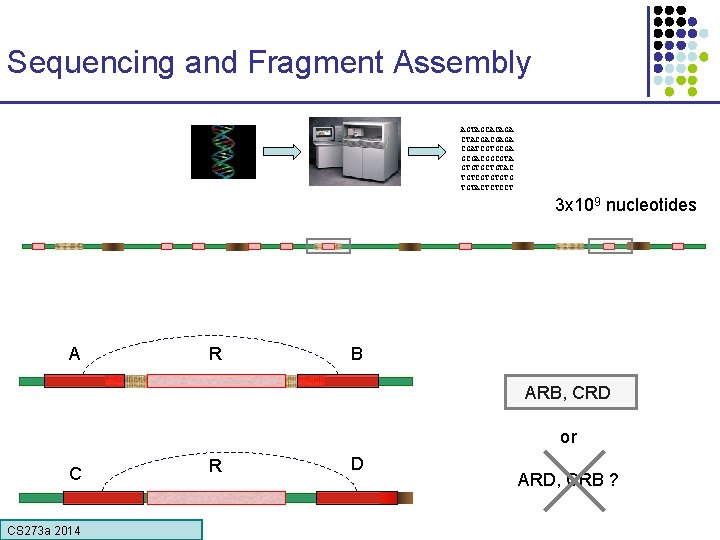
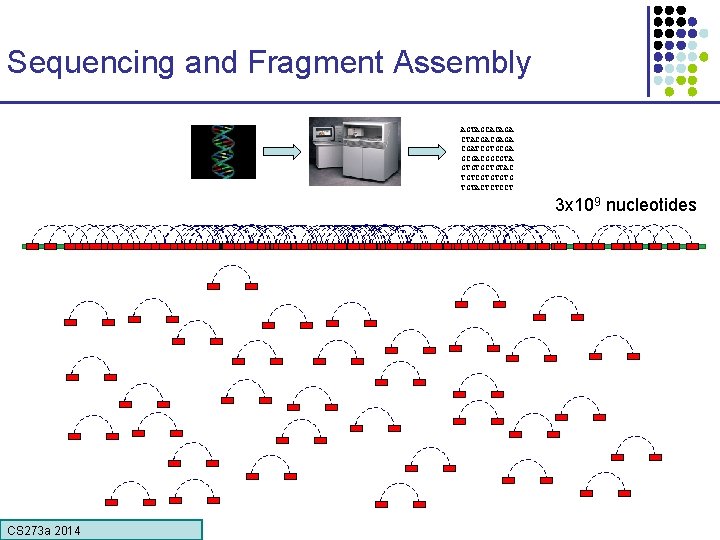
Sequencing and Fragment Assembly AGTAGCACAGA CTACGACGAGA CGATCGTGCGACGGCGTA GTGTGCTGTAC TGTCGTGTGTG TGTACTCTCCT 3 x 109 nucleotides 50% of human DNA is composed of repeats Error! Glued together two distant regions CS 273 a Lecture 4, Autumn 08, Batzoglou CS 273 a 2014

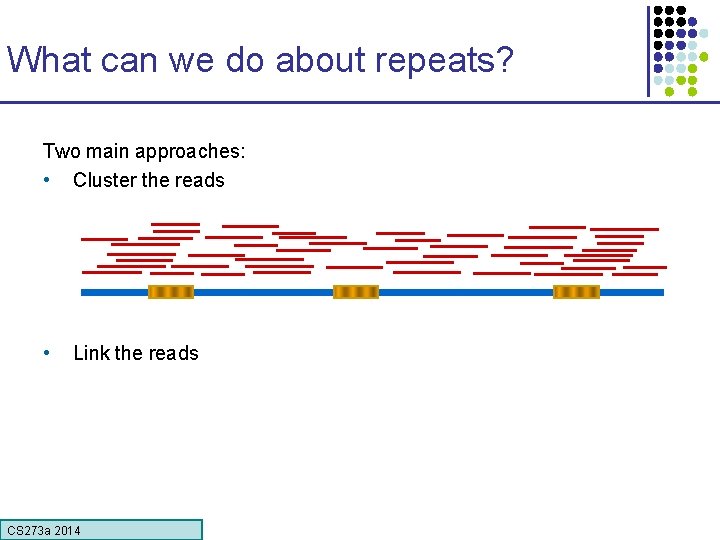
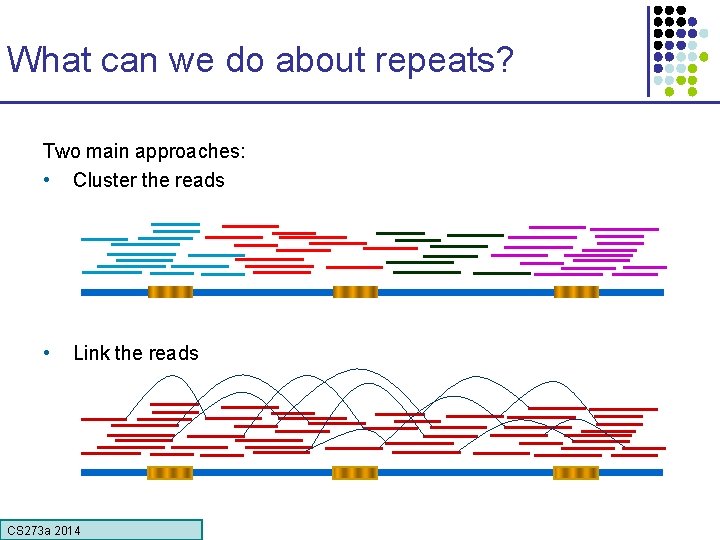
What can we do about repeats? Two main approaches: • Cluster the reads • Link the reads CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

What can we do about repeats? Two main approaches: • Cluster the reads • Link the reads CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

What can we do about repeats? Two main approaches: • Cluster the reads • Link the reads CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

Sequencing and Fragment Assembly AGTAGCACAGA CTACGACGAGA CGATCGTGCGACGGCGTA GTGTGCTGTAC TGTCGTGTGTG TGTACTCTCCT 3 x 109 nucleotides A R B ARB, CRD or C CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou R D ARD, CRB ?

Sequencing and Fragment Assembly AGTAGCACAGA CTACGACGAGA CGATCGTGCGACGGCGTA GTGTGCTGTAC TGTCGTGTGTG TGTACTCTCCT 3 x 109 nucleotides CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

Long Reads The Holy Grail CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

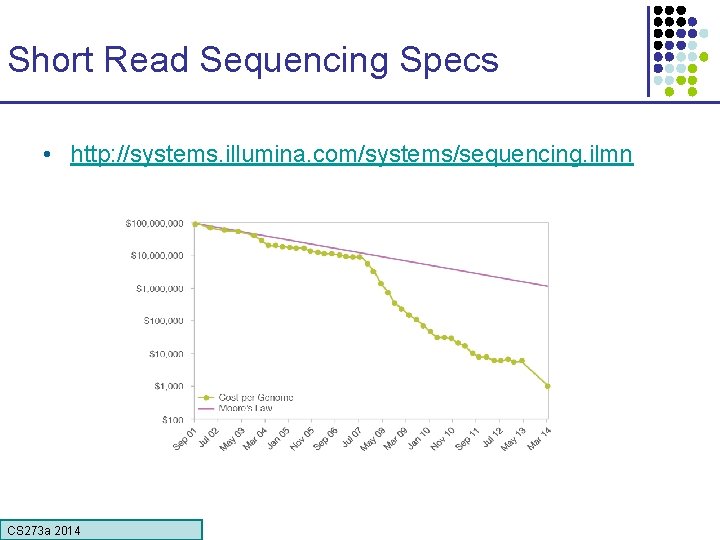
Short Read Sequencing Specs • http: //systems. illumina. com/systems/sequencing. ilmn CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

Long Reads - Pac. Bio Chemistry RS II: P 4 -C 2 RS II: P 5 -C 3 Optimized For higher quality longer reads Run time 180 min Total output ~275 Mb ~375 Mb Output/day ~2. 2 Gb ~3 Gb Mean read length ~5. 5 kb ~8. 5 kb Top 5% of reads >11 kb >18 kb Single pass accuracy ~86% ~83% >99. 999% >99. 98% ~50 k Instrument price ~$700 k Run price ~$400 Consensus (50 X) accuracy # of reads CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

Long Reads – Oxford Nanopore Read length: 50, 000+? Cost ? CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

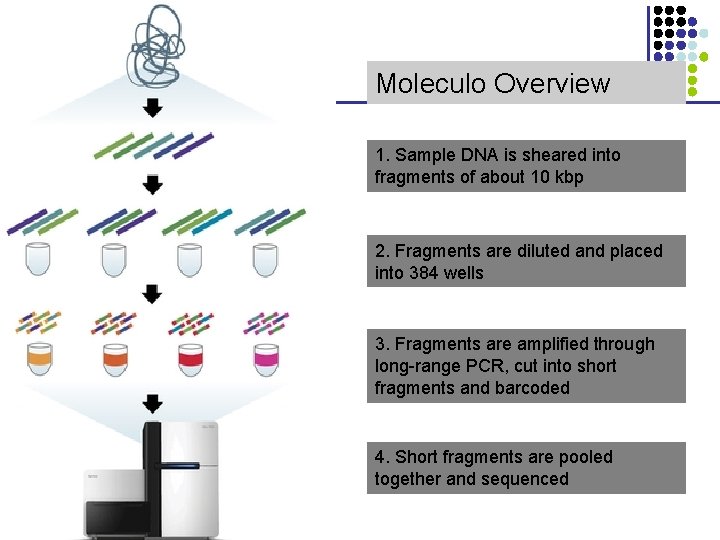
Moleculo Overview 1. Sample DNA is sheared into fragments of about 10 kbp 2. Fragments are diluted and placed into 384 wells 3. Fragments are amplified through long-range PCR, cut into short fragments and barcoded 4. Short fragments are pooled together and sequenced CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

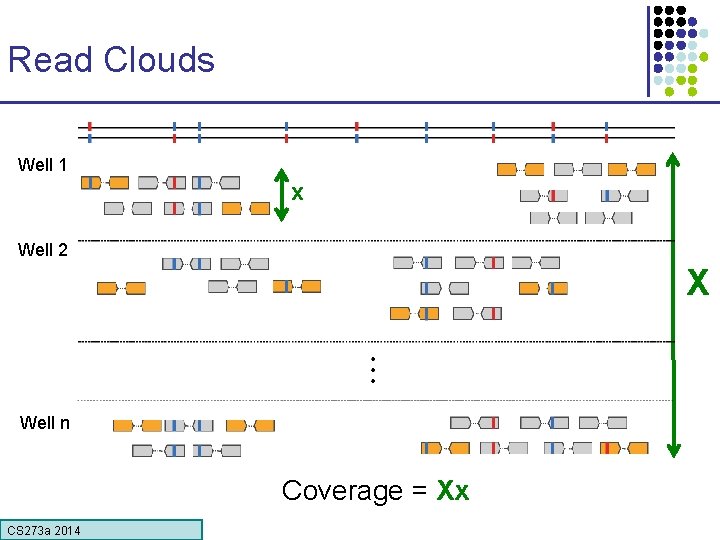
Read Clouds Well 1 x Well 2 X Well n Coverage = Xx CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

Fragment Assembly (in whole-genome shotgun sequencing) CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

Fragment Assembly Given N reads… Where N ~ 30 million… We need to use a linear-time algorithm CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

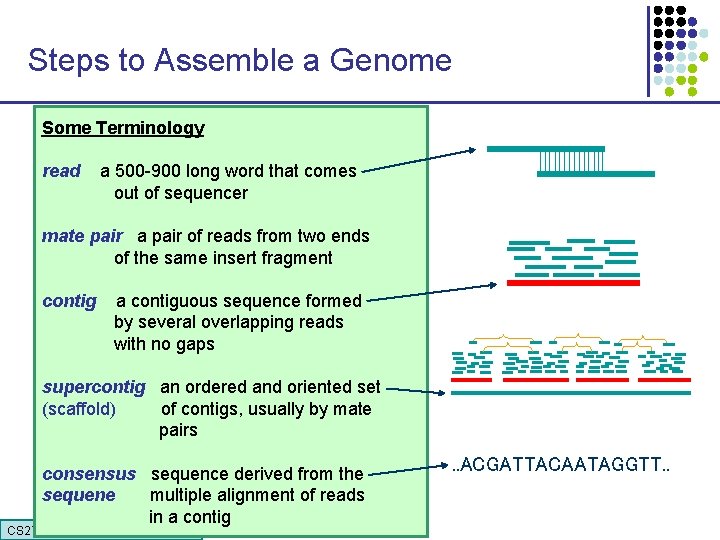
Steps to Assemble a Genome Some Terminology 1. Find overlapping readsthat comes read a 500 -900 long word out of sequencer mate pair a pair of reads from two ends 2. Merge some “good” of reads into of the same insert pairs fragment longer contigs contig a contiguous sequence formed by several overlapping reads with no gaps 3. Link contigs to form supercontigs supercontig an ordered and oriented set (scaffold) of contigs, usually by mate pairs 4. Derive consensus sequence derived from the sequene multiple alignment of reads in a contig CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou . . ACGATTACAATAGGTT. .

1. Find Overlapping Reads aaactgcagtacggatct aaactgcagt … gtacggatct gggcccaaactgcagtac gggcccaaac … actgcagtac gtacggatctactacaca gtacggatct … ctactacaca CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou (read, pos. , word, orient. ) (word, read, orient. , pos. ) aaactgcagt actgcagta … gtacggatct gggcccaaac gcccaaact … actgcagtac gtacggatct acggatcta … ctactacaca aaactgcagt acggatcta actgcagta cccaaactg cggatctactacac ctgcagtac gcccaaact ggcccaaac gggcccaaa gtacggatct tactacaca

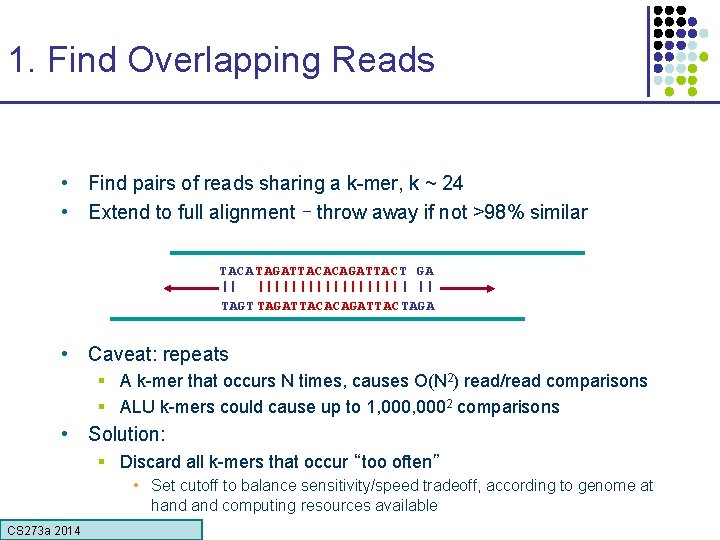
1. Find Overlapping Reads • Find pairs of reads sharing a k-mer, k ~ 24 • Extend to full alignment – throw away if not >98% similar TACA TAGATTACACAGATTAC T GA || ||||||||| | || TAGT TAGATTACACAGATTAC TAGA • Caveat: repeats § A k-mer that occurs N times, causes O(N 2) read/read comparisons § ALU k-mers could cause up to 1, 0002 comparisons • Solution: § Discard all k-mers that occur “too often” • Set cutoff to balance sensitivity/speed tradeoff, according to genome at hand computing resources available CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

1. Find Overlapping Reads Create local multiple alignments from the overlapping reads TAGATTACACAGATTACTGA TAG TTACACAGATTATTGA TAGATTACACAGATTACTGA CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

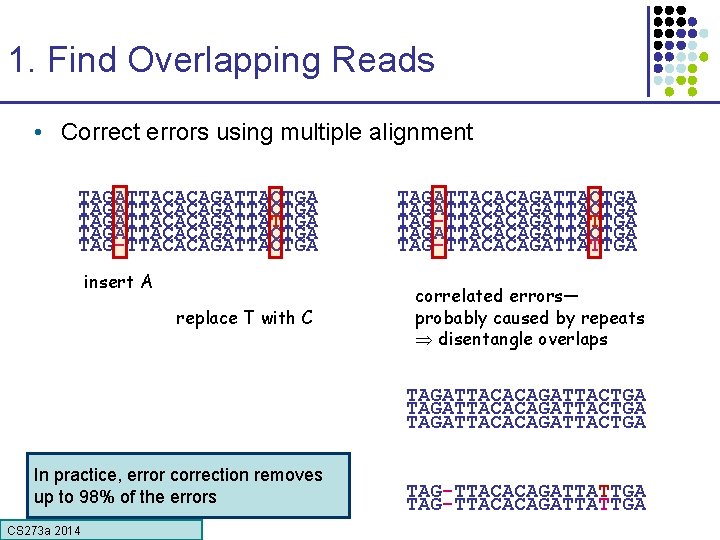
1. Find Overlapping Reads • Correct errors using multiple alignment TAGATTACACAGATTACTGA TAGATTACACAGATTATTGA TAGATTACACAGATTACTGA TAG-TTACACAGATTACTGA insert A replace T with C TAGATTACACAGATTACTGA TAG-TTACACAGATTATTGA correlated errors— probably caused by repeats disentangle overlaps TAGATTACACAGATTACTGA In practice, error correction removes up to 98% of the errors CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou TAG-TTACACAGATTATTGA

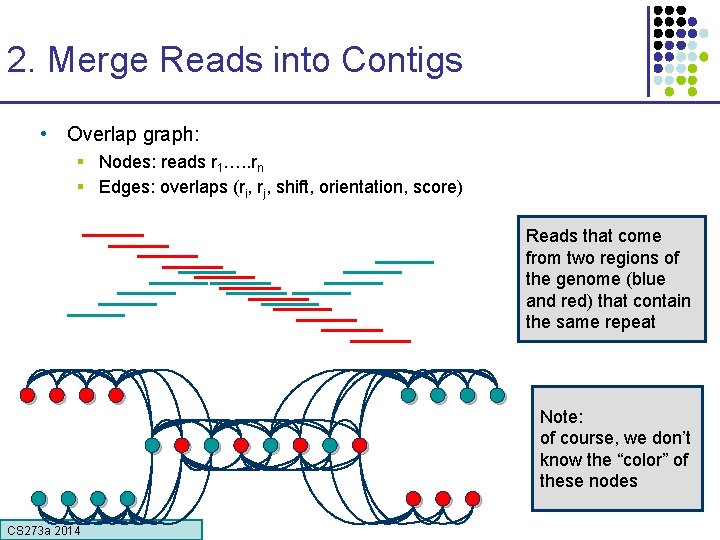
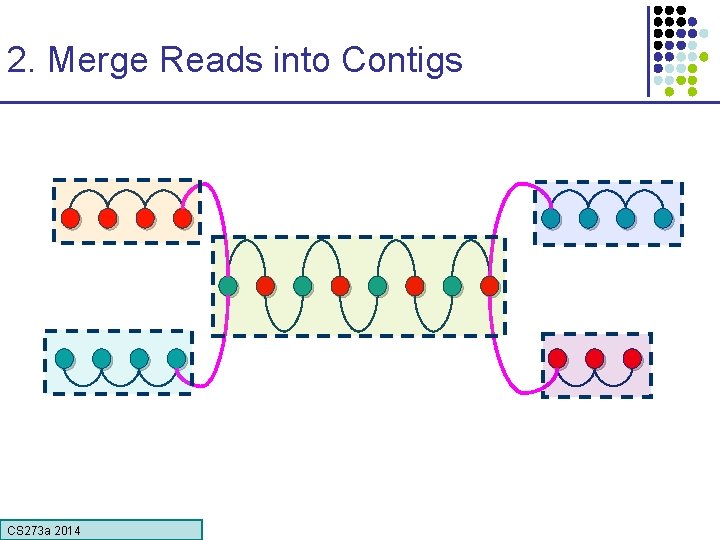
2. Merge Reads into Contigs • Overlap graph: § Nodes: reads r 1…. . rn § Edges: overlaps (ri, rj, shift, orientation, score) Reads that come from two regions of the genome (blue and red) that contain the same repeat Note: of course, we don’t know the “color” of these nodes CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

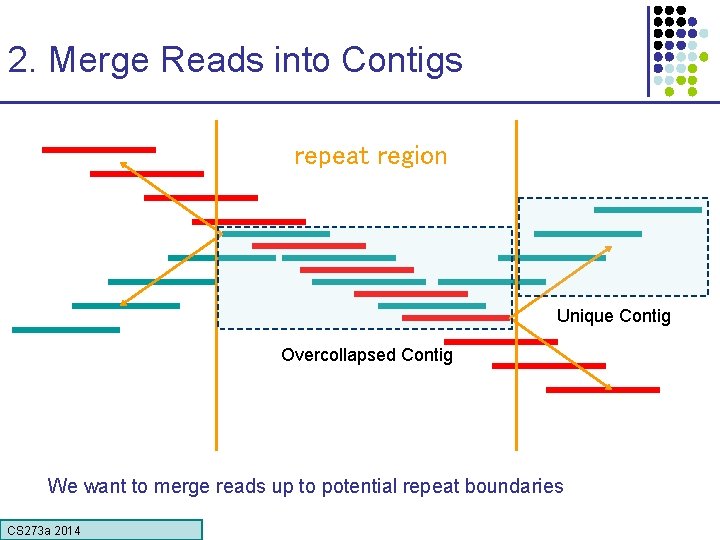
2. Merge Reads into Contigs repeat region Unique Contig Overcollapsed Contig We want to merge reads up to potential repeat boundaries CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

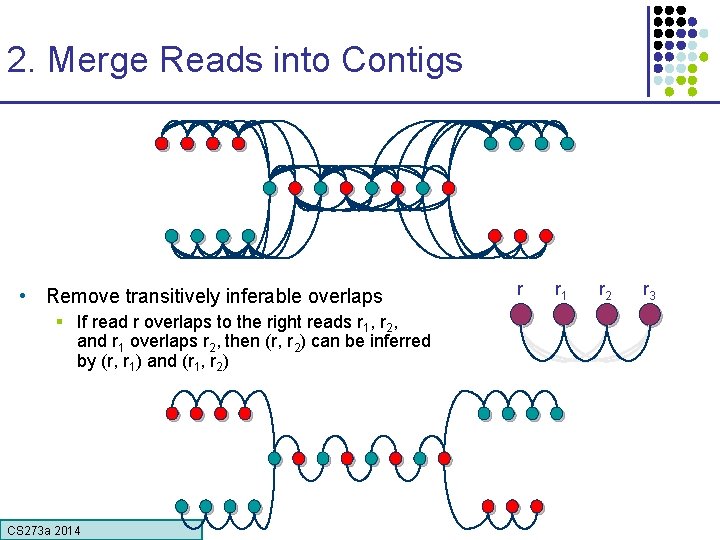
2. Merge Reads into Contigs • Remove transitively inferable overlaps § If read r overlaps to the right reads r 1, r 2, and r 1 overlaps r 2, then (r, r 2) can be inferred by (r, r 1) and (r 1, r 2) CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou r r 1 r 2 r 3

2. Merge Reads into Contigs CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

Repeats, errors, and contig lengths • Repeats shorter than read length are easily resolved § Read that spans across a repeat disambiguates order of flanking regions • Repeats with more base pair diffs than sequencing error rate are OK § We throw overlaps between two reads in different copies of the repeat • To make the genome appear less repetitive, try to: § Increase read length § Decrease sequencing error rate Role of error correction: Discards up to 98% of single-letter sequencing errors decreases error rate decreases effective repeat content increases contig length CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

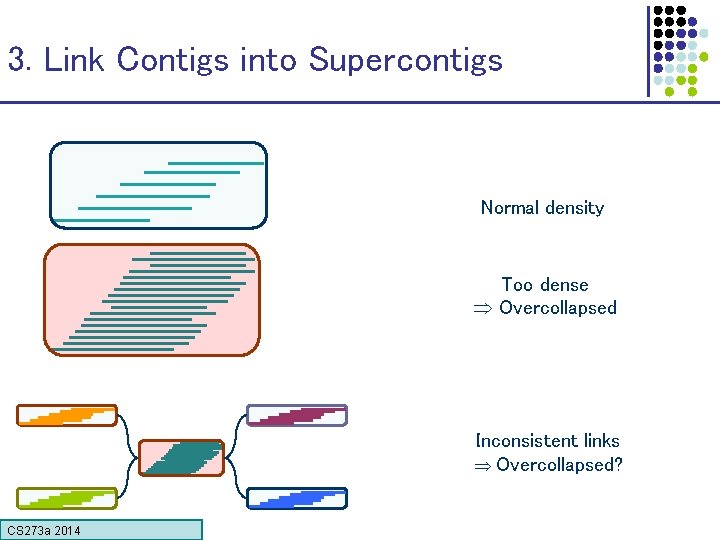
3. Link Contigs into Supercontigs Normal density Too dense Overcollapsed Inconsistent links Overcollapsed? CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

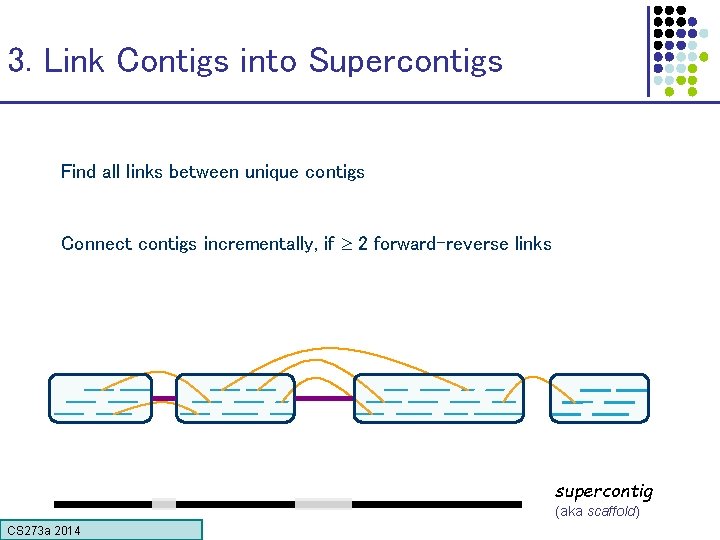
3. Link Contigs into Supercontigs Find all links between unique contigs Connect contigs incrementally, if 2 forward-reverse links supercontig (aka scaffold) CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

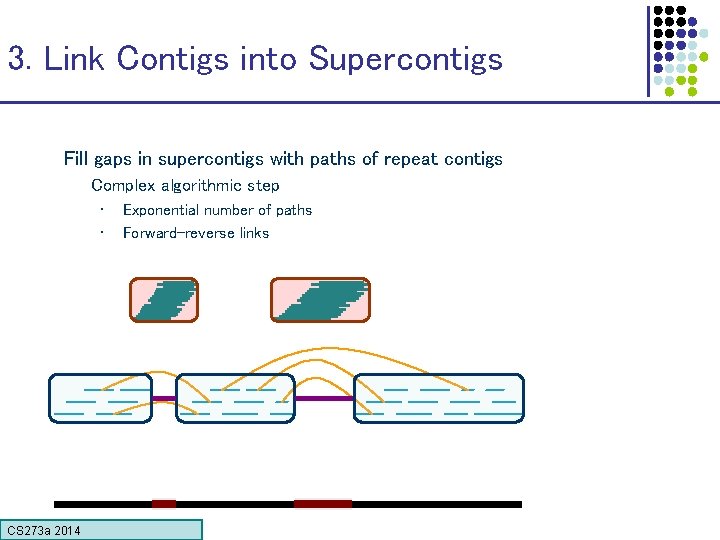
3. Link Contigs into Supercontigs Fill gaps in supercontigs with paths of repeat contigs Complex algorithmic step • • Exponential number of paths Forward-reverse links CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

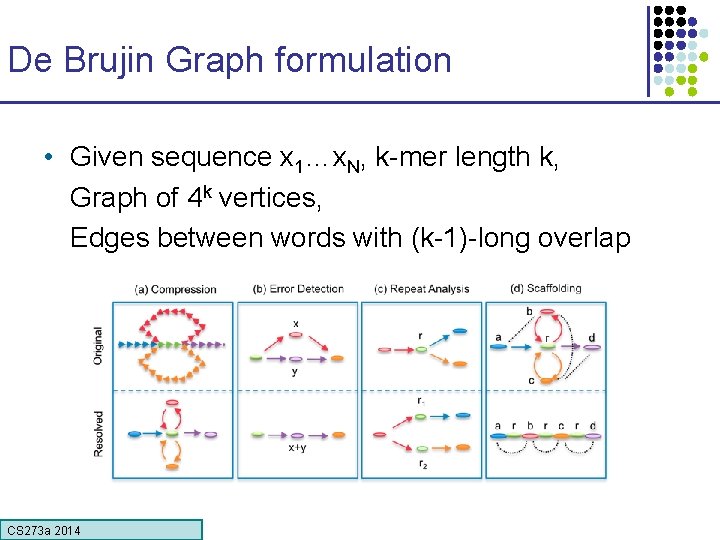
De Brujin Graph formulation • Given sequence x 1…x. N, k-mer length k, Graph of 4 k vertices, Edges between words with (k-1)-long overlap CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

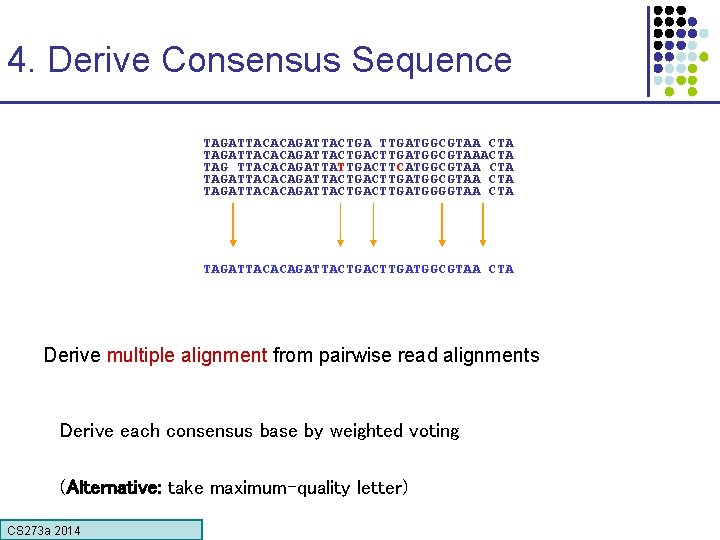
4. Derive Consensus Sequence TAGATTACACAGATTACTGA TTGATGGCGTAA CTA TAGATTACACAGATTACTGACTTGATGGCGTAAACTA TAG TTACACAGATTATTGACTTCATGGCGTAA CTA TAGATTACACAGATTACTGACTTGATGGGGTAA CTA TAGATTACACAGATTACTGACTTGATGGCGTAA CTA Derive multiple alignment from pairwise read alignments Derive each consensus base by weighted voting (Alternative: take maximum-quality letter) CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

Panda Genome CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou

History of WGA 1997 • 1982: -virus, 48, 502 bp • 1995: h-influenzae, Let’s sequence the human 1 genome Mbp with the shotgun strategy • 2000: fly, 100 Mbp • 2001 – present Thatrat is*, chicken, dog, chimpanzee, § human (3 Gbp), mouse (2. 5 Gbp), several fungal genomes impossible, and a bad idea anyway Gene Myers CS 273 a Lecture CS 273 a 2014 4, Autumn 08, Batzoglou Phil Green