Finding approximate palindromes in genomic sequences Project goals







- Slides: 12

Finding approximate palindromes in genomic sequences.

Project goals n Implementation of an algorithm for finding approximate palindromes in genomic sequences. n Usage of the algorithm for purposes of creating “palindrome fingerprints”. n Develop methods for testing the significance of specific approximate palindromes.

Background n Palindrome - A double strand DNA locus whose 5'-to-3' sequence is identical on each DNA strand. The sequence is the same when one strand is read left to right and the other strand is read right to the left. n Alternatively looking on one strand of the DNA the definition for palindrome is: A region of sequence, that when it’s been read left to right it is complementary to the sequence that been read right to left (A match T, and C match G).

Approximate Palindrome contain a certain number of mismatches and allow gap. “palindrome fingerprints” -Each DNA sequence has it’s unique number, sizes of palindromes, and location in sequence.

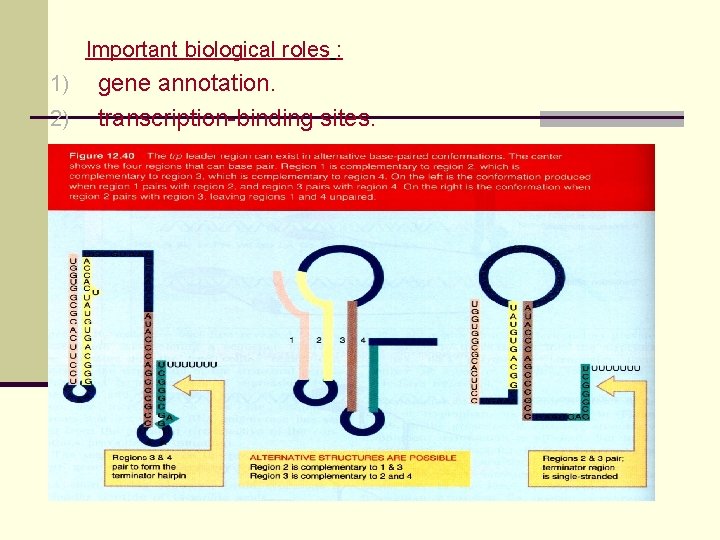
Important biological roles : 1) 2) gene annotation. transcription-binding sites.

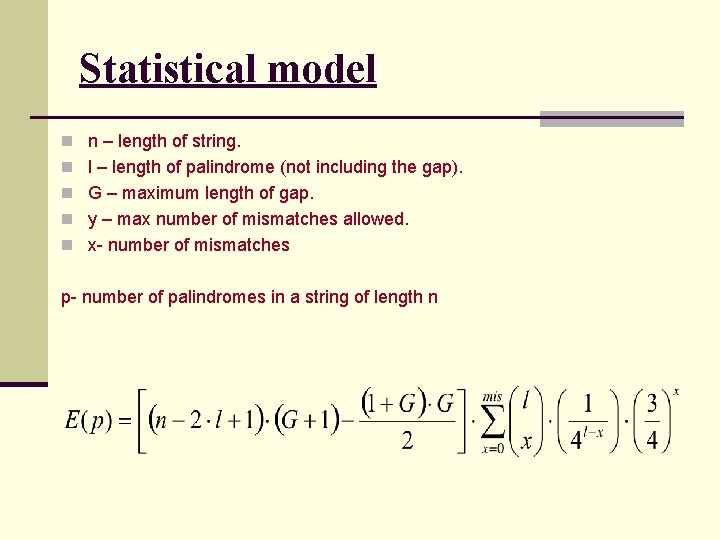
Statistical model n n – length of string. n l – length of palindrome (not including the gap). n G – maximum length of gap. n y – max number of mismatches allowed. n x- number of mismatches p- number of palindromes in a string of length n

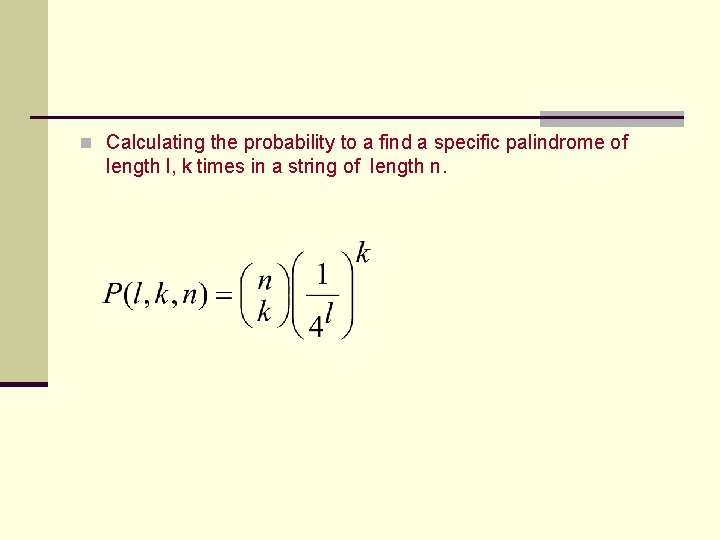
n Calculating the probability to a find a specific palindrome of length l, k times in a string of length n.

Application n Implemented in C n Input: 1) Sequence, genome of different organisms, text file in a FASTA format. 2) Length of palindrome (one side). 3) Maximum gap between repeated regions. 4) Number of mismatches allowed. n Output - all the palindromes within a specified length range and also a range of mismatch.

The Algorithm: > Search for the palindrome within a “window”, in the size of Max. Size. > Each iteration incrementing the size of palindrome, until Max. Size is reached. > Shift left of the window.

Algorithm Testing “Plant” an approximate palindrome in different genomes and compare the results with our expectations. Compare our formula expectation with the result of several random sequences

Practical usage of the Algorithm Compare the palindrome profile of different organisms and evaluate the results: n n n Genome from different bacteria. Same gene, for example: hemoglobin, insulin in different mammals. Gene families, for example: Histones, Immunoglobins.

n The End.