Figure 1 Schematic of picornavirus genome organization The
- Slides: 11

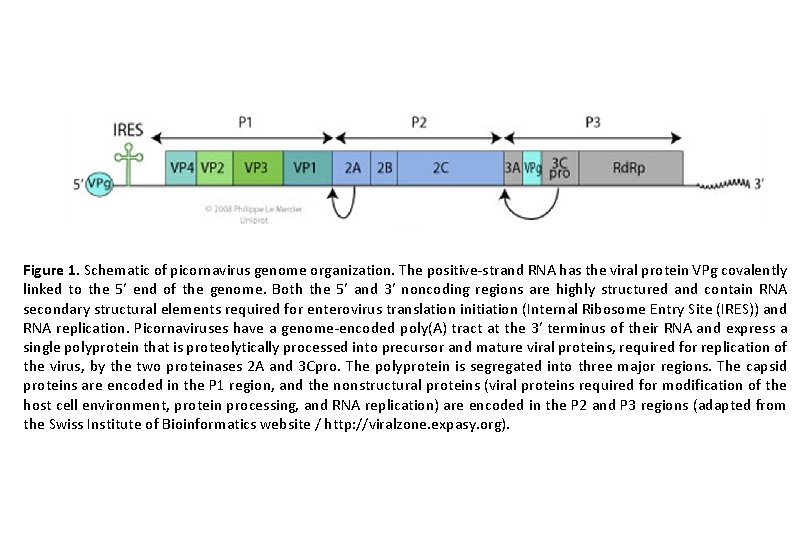
Figure 1. Schematic of picornavirus genome organization. The positive-strand RNA has the viral protein VPg covalently linked to the 5′ end of the genome. Both the 5′ and 3′ noncoding regions are highly structured and contain RNA secondary structural elements required for enterovirus translation initiation (Internal Ribosome Entry Site (IRES)) and RNA replication. Picornaviruses have a genome-encoded poly(A) tract at the 3′ terminus of their RNA and express a single polyprotein that is proteolytically processed into precursor and mature viral proteins, required for replication of the virus, by the two proteinases 2 A and 3 Cpro. The polyprotein is segregated into three major regions. The capsid proteins are encoded in the P 1 region, and the nonstructural proteins (viral proteins required for modification of the host cell environment, protein processing, and RNA replication) are encoded in the P 2 and P 3 regions (adapted from the Swiss Institute of Bioinformatics website / http: //viralzone. expasy. org).

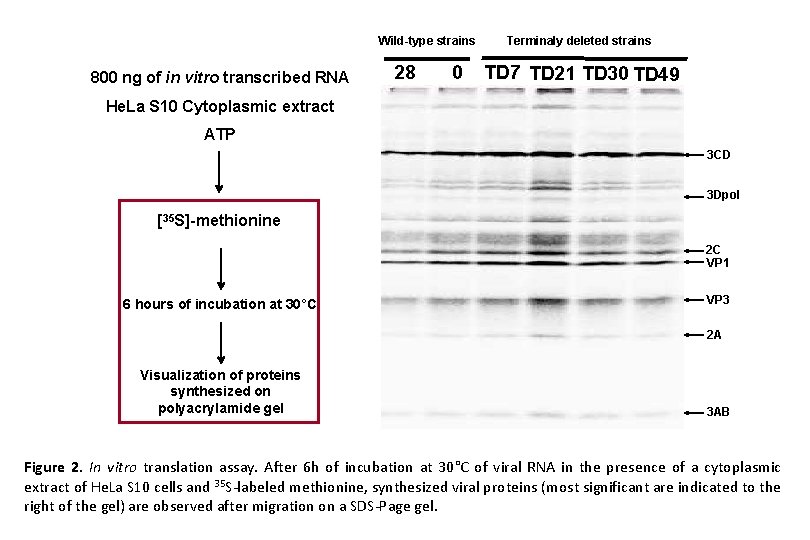
Wild-type strains 800 ng of in vitro transcribed RNA 28 0 Terminaly deleted strains TD 7 TD 21 TD 30 TD 49 He. La S 10 Cytoplasmic extract ATP 3 CD 3 Dpol [35 S]-methionine 2 C VP 1 6 hours of incubation at 30°C VP 3 2 A Visualization of proteins synthesized on polyacrylamide gel 3 AB Figure 2. In vitro translation assay. After 6 h of incubation at 30°C of viral RNA in the presence of a cytoplasmic extract of He. La S 10 cells and 35 S-labeled methionine, synthesized viral proteins (most significant are indicated to the right of the gel) are observed after migration on a SDS-Page gel.

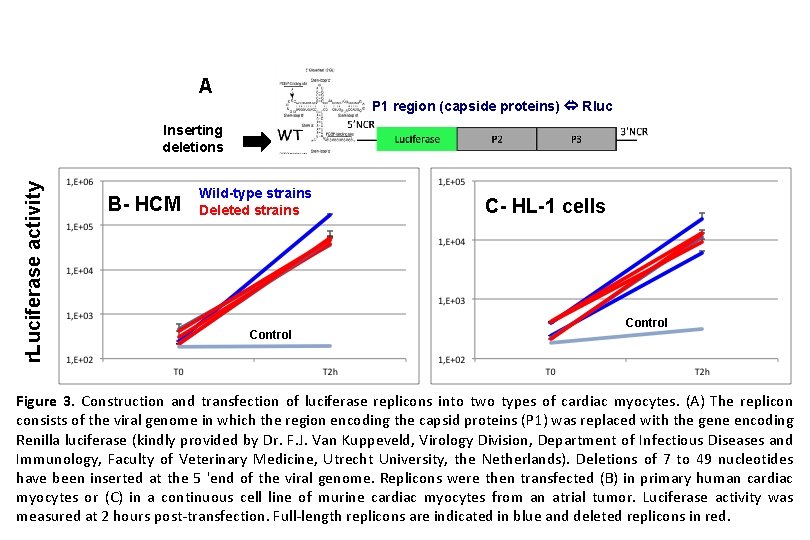
A P 1 region (capside proteins) Rluc r. Luciferase activity Inserting deletions B- HCM Wild-type strains Deleted strains Control C- HL-1 cells Control Figure 3. Construction and transfection of luciferase replicons into two types of cardiac myocytes. (A) The replicon consists of the viral genome in which the region encoding the capsid proteins (P 1) was replaced with the gene encoding Renilla luciferase (kindly provided by Dr. F. J. Van Kuppeveld, Virology Division, Department of Infectious Diseases and Immunology, Faculty of Veterinary Medicine, Utrecht University, the Netherlands). Deletions of 7 to 49 nucleotides have been inserted at the 5 'end of the viral genome. Replicons were then transfected (B) in primary human cardiac myocytes or (C) in a continuous cell line of murine cardiac myocytes from an atrial tumor. Luciferase activity was measured at 2 hours post-transfection. Full-length replicons are indicated in blue and deleted replicons in red.

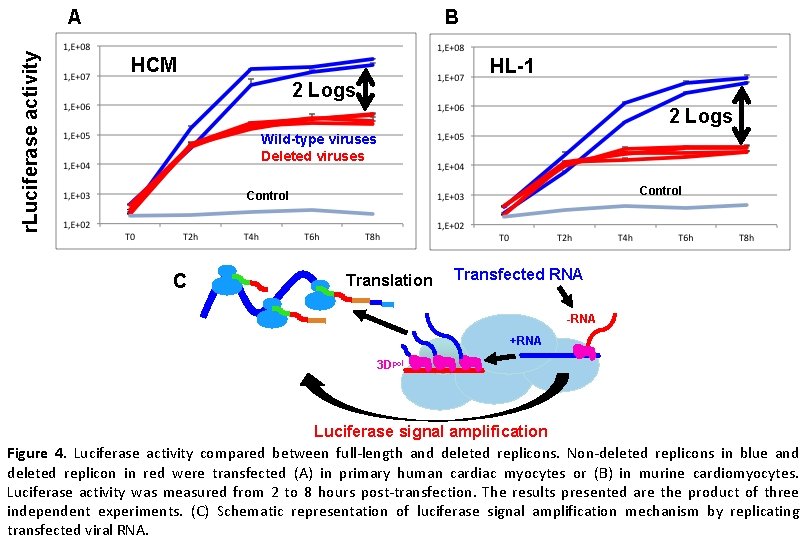
r. Luciferase activity A B HCM HL-1 2 Logs Wild-type viruses Deleted viruses Control C Translation Transfected RNA -RNA +RNA 3 Dpol Luciferase signal amplification Figure 4. Luciferase activity compared between full-length and deleted replicons. Non-deleted replicons in blue and deleted replicon in red were transfected (A) in primary human cardiac myocytes or (B) in murine cardiomyocytes. Luciferase activity was measured from 2 to 8 hours post-transfection. The results presented are the product of three independent experiments. (C) Schematic representation of luciferase signal amplification mechanism by replicating transfected viral RNA.

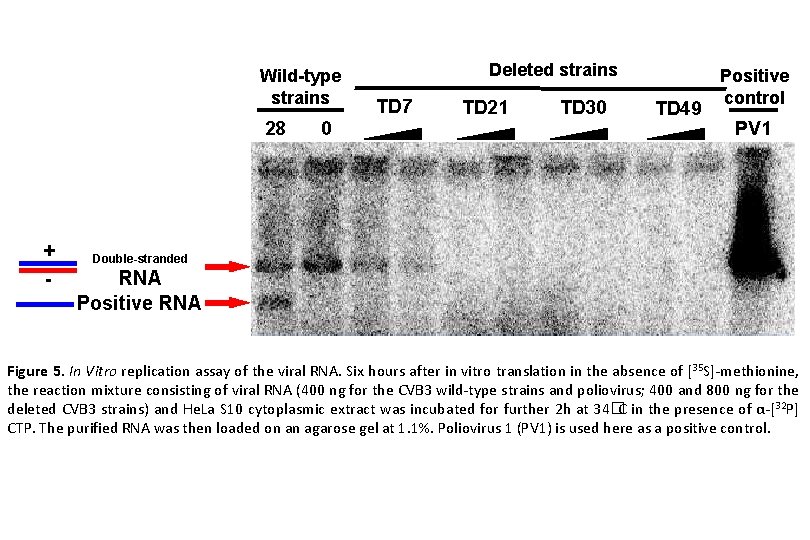
Wild-type strains 28 + - 0 Deleted strains TD 7 TD 21 TD 30 TD 49 Positive control PV 1 Double-stranded RNA Positive RNA Figure 5. In Vitro replication assay of the viral RNA. Six hours after in vitro translation in the absence of [35 S]-methionine, the reaction mixture consisting of viral RNA (400 ng for the CVB 3 wild-type strains and poliovirus; 400 and 800 ng for the deleted CVB 3 strains) and He. La S 10 cytoplasmic extract was incubated for further 2 h at 34�C in the presence of α-[32 P] CTP. The purified RNA was then loaded on an agarose gel at 1. 1%. Poliovirus 1 (PV 1) is used here as a positive control.

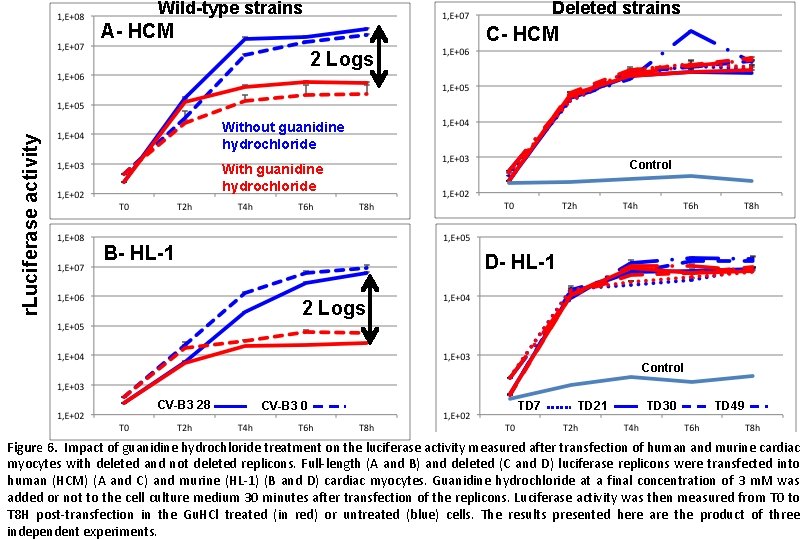
Wild-type strains Deleted strains A- HCM C- HCM r. Luciferase activity 2 Logs Without guanidine hydrochloride Control With guanidine hydrochloride B- HL-1 D- HL-1 2 Logs Control CV-B 3 28 CV-B 3 0 TD 7 TD 21 TD 30 TD 49 Figure 6. Impact of guanidine hydrochloride treatment on the luciferase activity measured after transfection of human and murine cardiac myocytes with deleted and not deleted replicons. Full-length (A and B) and deleted (C and D) luciferase replicons were transfected into human (HCM) (A and C) and murine (HL-1) (B and D) cardiac myocytes. Guanidine hydrochloride at a final concentration of 3 m. M was added or not to the cell culture medium 30 minutes after transfection of the replicons. Luciferase activity was then measured from T 0 to T 8 H post-transfection in the Gu. HCl treated (in red) or untreated (blue) cells. The results presented here are the product of three independent experiments.

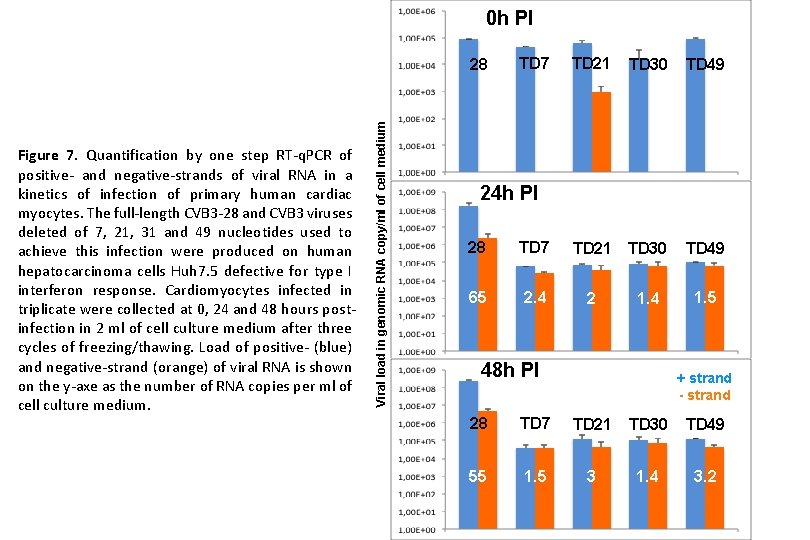
0 h PI Figure 7. Quantification by one step RT-q. PCR of positive- and negative-strands of viral RNA in a kinetics of infection of primary human cardiac myocytes. The full-length CVB 3 -28 and CVB 3 viruses deleted of 7, 21, 31 and 49 nucleotides used to achieve this infection were produced on human hepatocarcinoma cells Huh 7. 5 defective for type I interferon response. Cardiomyocytes infected in triplicate were collected at 0, 24 and 48 hours postinfection in 2 ml of cell culture medium after three cycles of freezing/thawing. Load of positive- (blue) and negative-strand (orange) of viral RNA is shown on the y-axe as the number of RNA copies per ml of cell culture medium. Viral load in genomic RNA copy/ml of cell medium 28 TD 7 TD 21 TD 30 TD 49 24 h PI 28 TD 7 65 2. 4 2 1. 4 48 h PI 1. 5 + strand - strand 28 TD 7 TD 21 TD 30 TD 49 55 1. 5 3 1. 4 3. 2

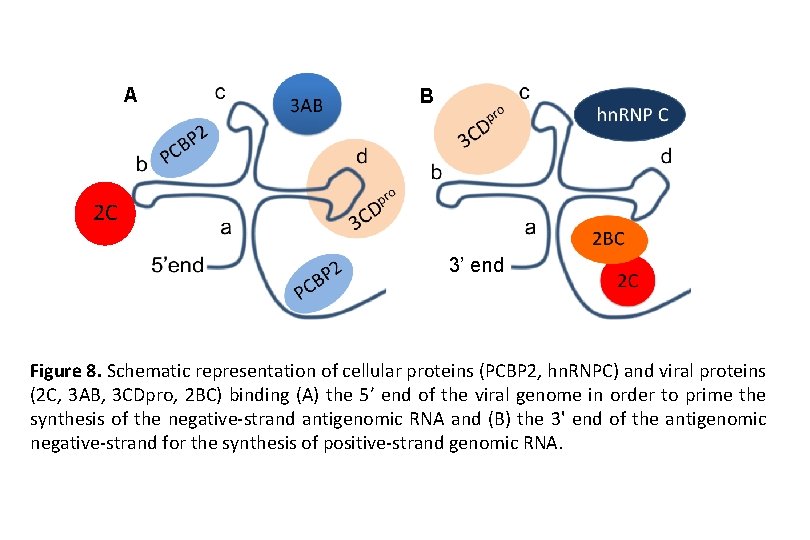
A B 2 C 3’ end Figure 8. Schematic representation of cellular proteins (PCBP 2, hn. RNPC) and viral proteins (2 C, 3 AB, 3 CDpro, 2 BC) binding (A) the 5’ end of the viral genome in order to prime the synthesis of the negative-strand antigenomic RNA and (B) the 3' end of the antigenomic negative-strand for the synthesis of positive-strand genomic RNA.

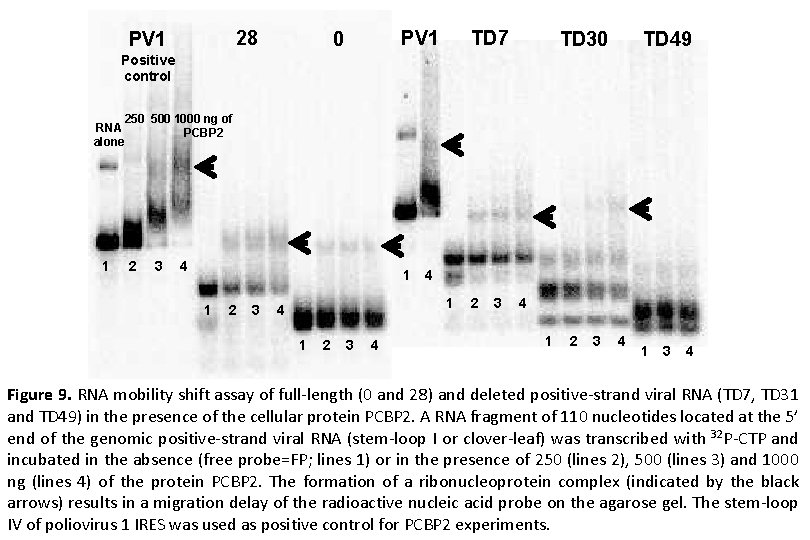
28 PV 1 0 TD 7 TD 30 TD 49 Positive control 250 500 1000 ng of RNA PCBP 2 alone 1 2 3 4 1 4 1 2 3 4 1 3 4 Figure 9. RNA mobility shift assay of full-length (0 and 28) and deleted positive-strand viral RNA (TD 7, TD 31 and TD 49) in the presence of the cellular protein PCBP 2. A RNA fragment of 110 nucleotides located at the 5’ end of the genomic positive-strand viral RNA (stem-loop I or clover-leaf) was transcribed with 32 P-CTP and incubated in the absence (free probe=FP; lines 1) or in the presence of 250 (lines 2), 500 (lines 3) and 1000 ng (lines 4) of the protein PCBP 2. The formation of a ribonucleoprotein complex (indicated by the black arrows) results in a migration delay of the radioactive nucleic acid probe on the agarose gel. The stem-loop IV of poliovirus 1 IRES was used as positive control for PCBP 2 experiments.

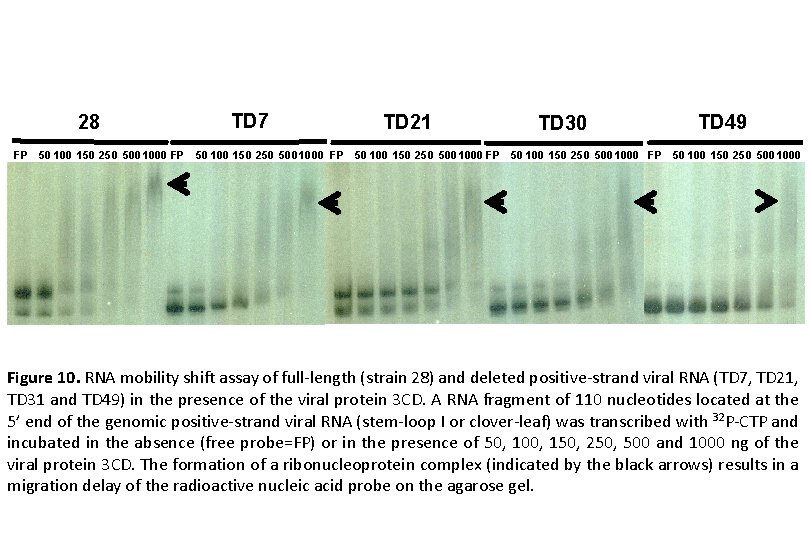
28 TD 7 TD 21 TD 30 TD 49 FP 50 100 150 250 500 1000 FP 50 100 150 250 500 1000 Figure 10. RNA mobility shift assay of full-length (strain 28) and deleted positive-strand viral RNA (TD 7, TD 21, TD 31 and TD 49) in the presence of the viral protein 3 CD. A RNA fragment of 110 nucleotides located at the 5’ end of the genomic positive-strand viral RNA (stem-loop I or clover-leaf) was transcribed with 32 P-CTP and incubated in the absence (free probe=FP) or in the presence of 50, 100, 150, 250, 500 and 1000 ng of the viral protein 3 CD. The formation of a ribonucleoprotein complex (indicated by the black arrows) results in a migration delay of the radioactive nucleic acid probe on the agarose gel.

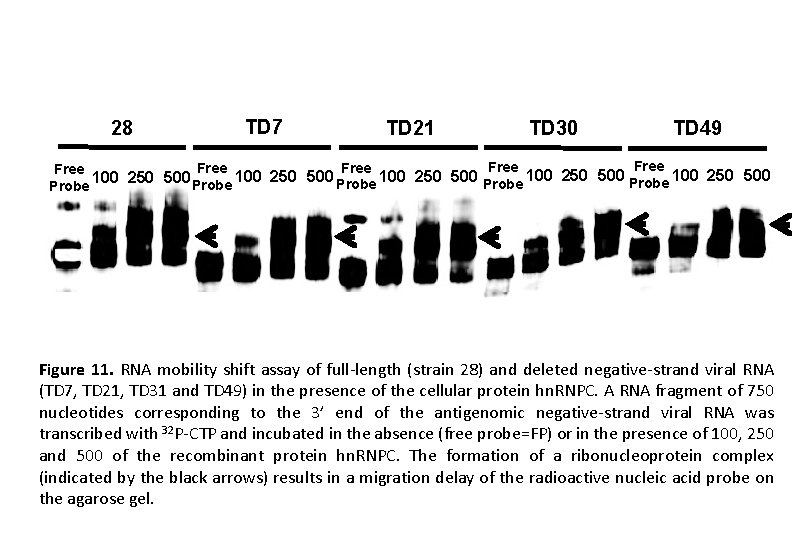
28 TD 7 TD 21 TD 30 TD 49 Free Free 100 250 500 100 250 500 Probe Probe Figure 11. RNA mobility shift assay of full-length (strain 28) and deleted negative-strand viral RNA (TD 7, TD 21, TD 31 and TD 49) in the presence of the cellular protein hn. RNPC. A RNA fragment of 750 nucleotides corresponding to the 3’ end of the antigenomic negative-strand viral RNA was transcribed with 32 P-CTP and incubated in the absence (free probe=FP) or in the presence of 100, 250 and 500 of the recombinant protein hn. RNPC. The formation of a ribonucleoprotein complex (indicated by the black arrows) results in a migration delay of the radioactive nucleic acid probe on the agarose gel.
 Semi-global alignment
Semi-global alignment Picornavirus familia
Picornavirus familia Contoh virus rna jenis picornaviridae
Contoh virus rna jenis picornaviridae Genome organization
Genome organization Process organization in computer organization
Process organization in computer organization Alternating pattern essay
Alternating pattern essay Patric bioinformatics
Patric bioinformatics Savant genome browser
Savant genome browser 14-3 human molecular genetics
14-3 human molecular genetics Prokaryotic gene structure
Prokaryotic gene structure Innovation genome project
Innovation genome project Difference between bac and yac
Difference between bac and yac