DNA Repair Copyright 2002 John Wiley Sons Inc



- Slides: 23

DNA Repair Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

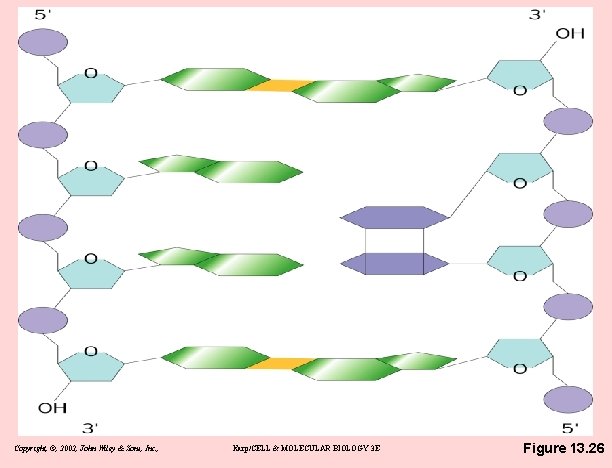
Stable, but fragile • Types of damage experience by DNA – Ionizing radiation can break DNA backbone – chemicals, some made by cell metabolism – ultraviolet radiation: pyrimidine dimers – thermal energy can depurinate adenine & guanine – warm-blooded mammals lose ~10, 000 bases/day Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E Figure 13. 26

Stable, but fragile • Failure to repair causes mutations – Can interfere with transcription and replication – Can lead to malignant transformation – Can speed aging • It is essential that cells possess mechanisms for repairing this damage – Repair mechanisms are extensive and efficient – <1 base change per thousand escapes repair Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Stable, but fragile • Many repair proteins – Repair is sometimes direct; but usually excised & replaced – One enzyme uses sunlight energy to fix pyrimidine dimers – Excision repair uses info in undamaged complementary strand – DNA replication & repair share many parts & services • Adverse effects seen in humans with repair defects Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

NER = Nucleotide Excision Repair • Works on bulky lesions like pyrimidine dimers & adducts • Uses "cut-and-patch" mechanism • 2 distinct NER pathways distinguished – transcription coupled pathway – slower global pathway Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

NER = Nucleotide Excision Repair • Transcription-coupled pathway – lesion detected by stalled RNA polymerase – transcribed genes are highest priority • Global pathway - slower, less efficient Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

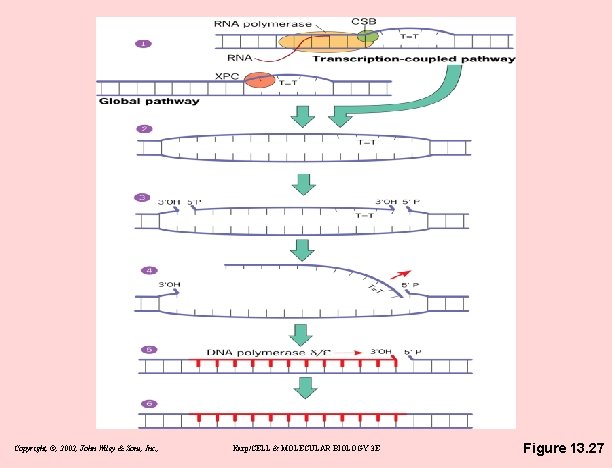
NER = Nucleotide Excision Repair • Damage recognition – 2 NER pathways differ in lesion recognition – subsequent repair steps are thought to be very similar – TFIIH (participates in transcription initiation, too) • A key component of repair machinery • link between transcription & DNA repair • two TFIIH subunits (XPB & XPD) are helicases • damaged strand released by endonuclease cleavage (about 30 bases) • gap filled by DNA polymerase, then ligase Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E Figure 13. 27

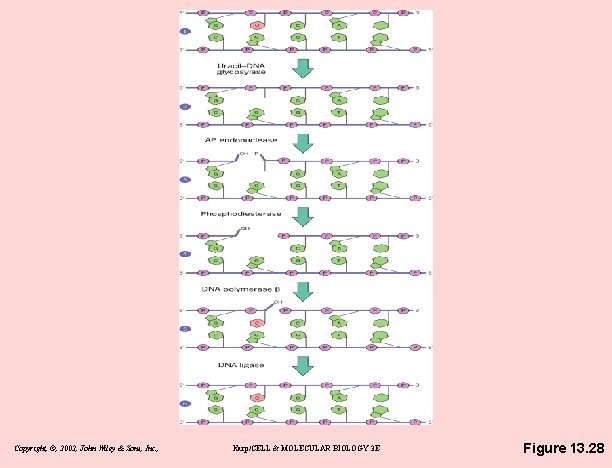
BER = Base Excision Repair • Base excision repair (BER) – remove damaged bases – alterations more subtle, distort the helix less – Steps of BER • DNA glycosylase removes base • cleaves glycosidic bond holding the base to sugar • "debased" deoxyribose phosphate removed • combined action of an endonuclease & a phosphodiesterase • Gap is then filled by DNA polymerase b & sealed by DNA ligase Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

BER = Base Excision Repair • Multiple DNA glycosylases – each is more-or-less specific for a type of altered base – Uracil - forms by hydrolytic removal of cytosine's amino group – 8 -hydroxyguanine - results from damage by oxygen free radicals – 3 -methyladenine - caused by alkylating agents Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

BER = Base Excision Repair • Uracil formation from cytosine – explains why thymine used instead of uracil – damage to cytosine = “normal” uracil – uracil-DNA glycosylase is highly conserved protein – E. coli & humans: 56% identity in amino acid sequence Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E Figure 13. 28

MMR = Mismatch Repair • enzyme removes mismatched nucleotide • in bacteria – Parental strand has methyl-adenosine residues – Provide signal for polarized repair – removes & replaces from nonmethylated strand – Returns correct base pair • in eukaryotes – the mechanism of identification of new strand unclear – does not appear to use methylation signal Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Double-strand breakage repair • Caused by ionizing radiation (X-rays, gamma rays) • Also caused by chemicals (bleomycin, free radicals) • Ultimately may prove lethal • DSBs can be repaired by several alternate pathways Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

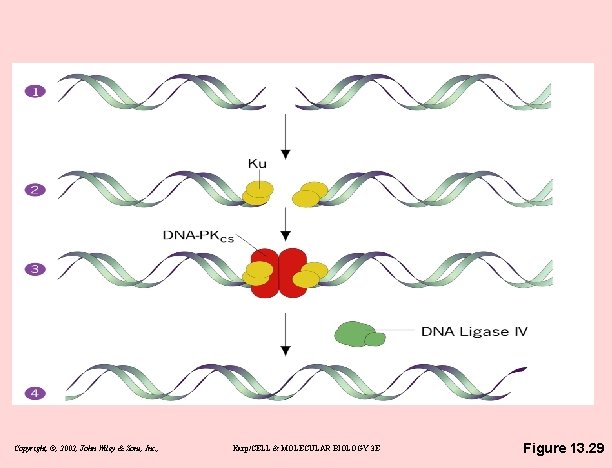
Double-strand breakage repair • NHEJ in mammalian cells – non-homologous end joining – the simplest & most commonly used – complex of proteins binds to broken ends – catalyzes a series of reactions that rejoin the broken strands – mutants for NHEJ are very sensitive to ionizing radiation Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E Figure 13. 29

Double-strand breakage repair • Another DSB repair pathway – includes genetic recombination – considerably more complex Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

DNA Replication and Repair Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Xeroderma pigmentosum (XP) • inherited disease • patients unable to repair damage from exposure to u. v. • defect in 1 of 7 different genes – nucleotide excision repair (NER) genes – XPA, XPB, XPC, XPD, XPE, XPF & XPG Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Xeroderma pigmentosum (XP) • patients susceptible to skin cancer via sun exposure – capable of nucleotide excision repair – only slightly more sensitive to UV light – but, produced fragmented daughter strands after UV irradiation – a variant form of XP, designated XP-V Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Unrepaired lesions block replication • Polymerase stalls • recruit specialized polymerase that is able to bypass the lesion – thymidine dimer as example – replicative polymerase (pol d or e) replaced pol h – This enzyme inserts 2 A residues across from dimer – XP-V mutation alters pol h – Cannot replicate past thymidine dimers Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E

Unrepaired lesions block replication • Polymerase h is member of a superfamily – bypass polymerases are “error prone” – trans-lesion synthesis (TLS) • different basic structure from classic DNA polymerases • they lack processivity: one or a few bases • no proofreading capability – humans have at least 30 TLS polymerases (genome project) Copyright, ©, 2002, John Wiley & Sons, Inc. , Karp/CELL & MOLECULAR BIOLOGY 3 E