Dali A Protein Structural Comparison Algorithm Using 2

- Slides: 13

Dali: A Protein Structural Comparison Algorithm Using 2 D Distance Matrices

Main Points for Discussion • Overview of why structural comparison can be a useful mode of analysis. • Using a 2 -D distance matrix to represent a 3 -D protein structure. • Specific computer algorithms that have been used to accomplish this analysis, including Monte Carlo optimization. • Further applications of Dali.

Why consider structural comparison? • 1 D sequence comparisons has traditionally been (and still is) used to determine degree of relatedness, although a low degree of sequence homology may yield surprisingly similar structures. • 3 D structural alignment is aimed at providing more information about the structure-function similarities between proteins with nondetectable evolutionary relationships.

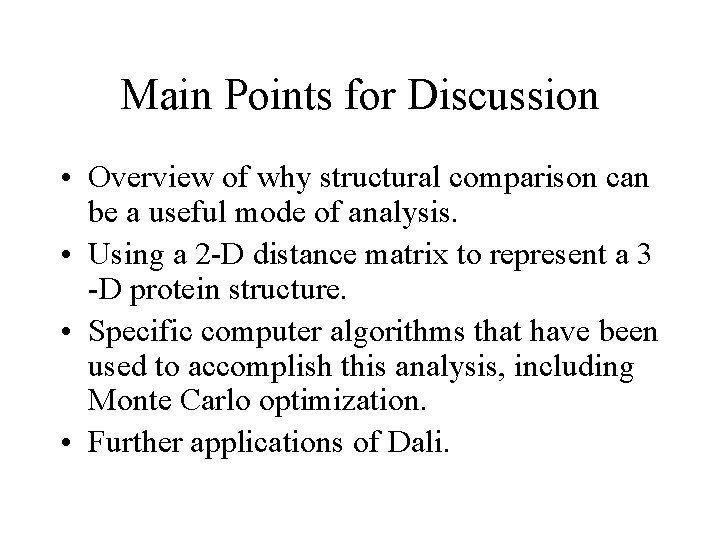
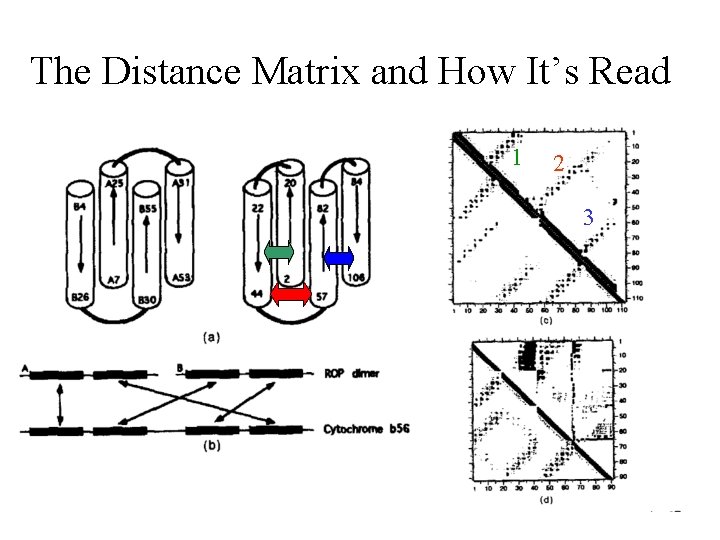
The Distance Matrix and How It’s Read 1 2 3

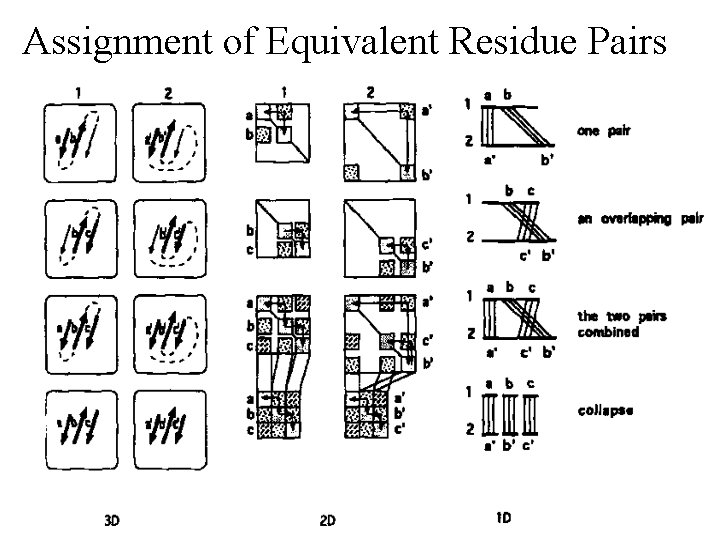
Assignment of Equivalent Residue Pairs

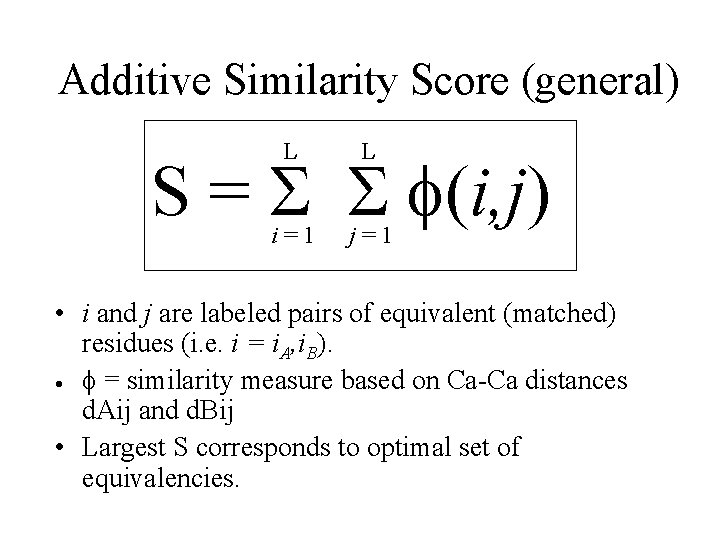
Additive Similarity Score (general) L L i=1 j=1 S = S S f(i, j) • i and j are labeled pairs of equivalent (matched) residues (i. e. i = i. A, i. B). · f = similarity measure based on Ca-Ca distances d. Aij and d. Bij • Largest S corresponds to optimal set of equivalencies.

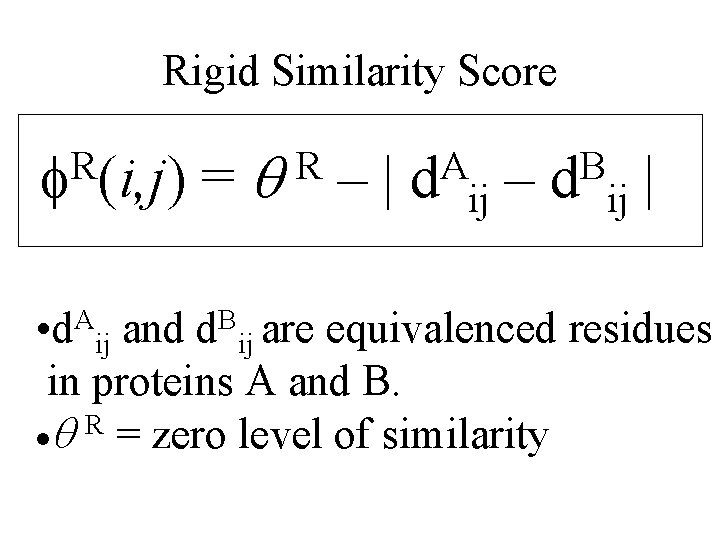
Rigid Similarity Score R f (i, j) =q –| R A d – ij B d | ij • d. Aij and d. Bij are equivalenced residues in proteins A and B. ·q R = zero level of similarity

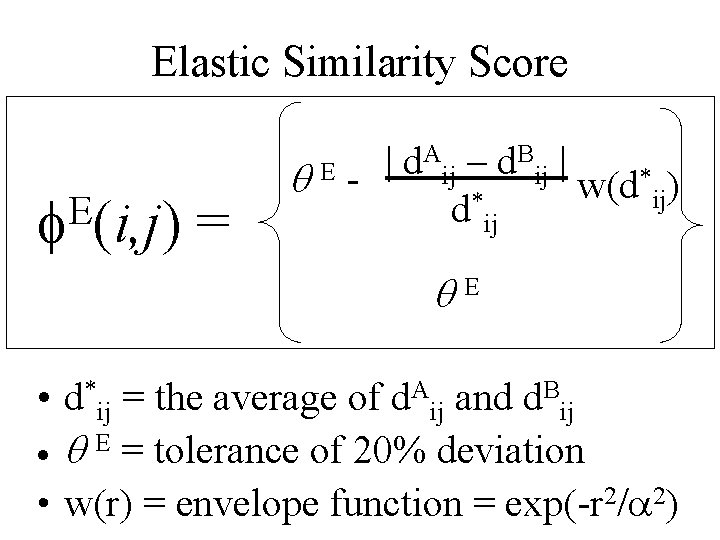
Elastic Similarity Score E f (i, j) = A – d. B | | d ij ij * ) q. Ew(d ij d*ij q. E • d*ij = the average of d. Aij and d. Bij · q E = tolerance of 20% deviation • w(r) = envelope function = exp(-r 2/a 2)

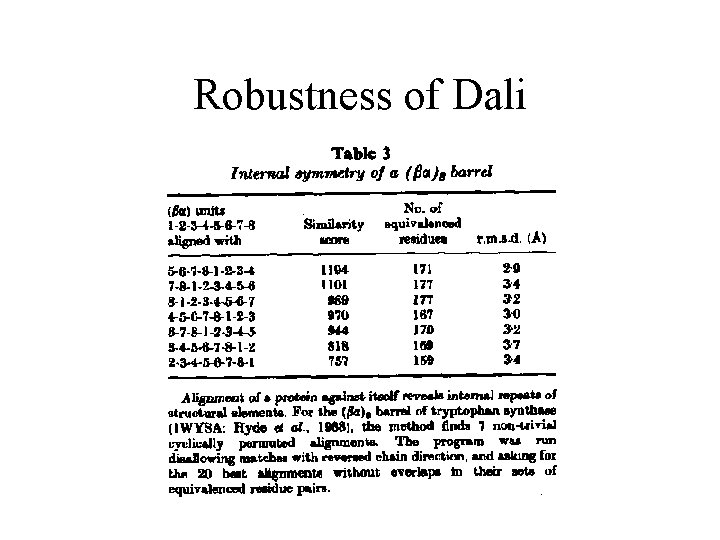
Robustness of Dali

Quality of Generated Alignments • Accuracy was verified by examining conserved functional residues in seeemingly divergent structures. • The elasticity score is useful in that it captures relative movements of structural elements (e. g. ATP binding site in hsp 70) and leaves only extremely non-homologous loops unaligned.

Quality of Generated Alignments (cont. ) • Detection of inter-domain motion brings functionally important residues into focus (e. g. ATP binding site in hsp 70). • Manipulation of the elastic similarity score determines the stringency of the alignment.

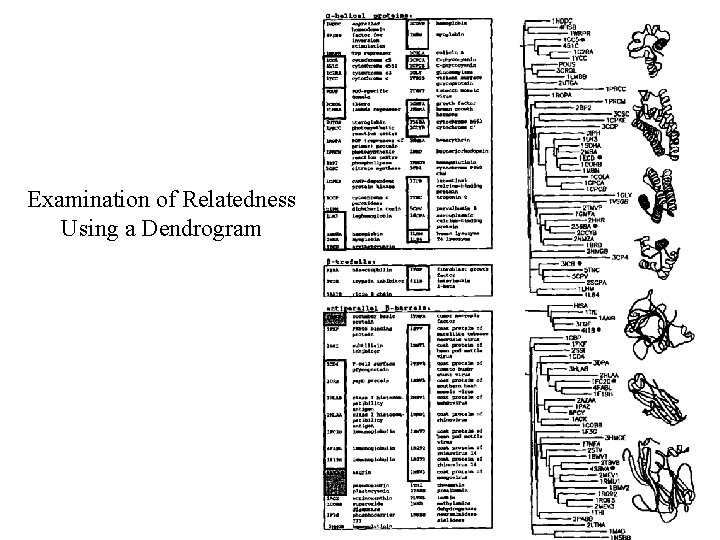
Dendrogram Examination of Relatedness Using a Dendrogram

Further Applications of Dali • Continuing further in an attempt to map the entire protein space using quantitative comparisons between structures (correspondence analysis on p. 133) • Applications to residue-residue energy interactions to create a more accurate biochemical representation of the protein. Also able to yield more useful information to predict 3 D structure from amino acid sequence due to the energies of interacting residues.