CSCI 2950 C Lecture 13 Network Motifs Network


- Slides: 22

CSCI 2950 -C Lecture 13 Network Motifs; Network Integration Ben Raphael November 20, 2008 http: //cs. brown. edu/courses/csci 2950 -c/

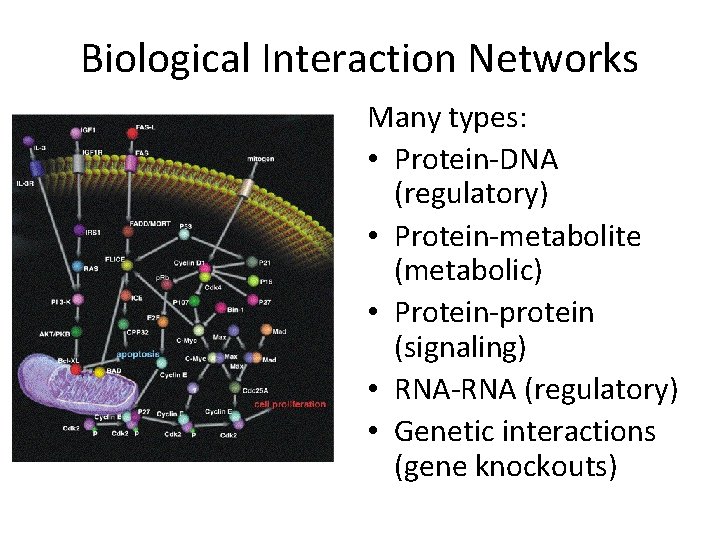
Biological Interaction Networks Many types: • Protein-DNA (regulatory) • Protein-metabolite (metabolic) • Protein-protein (signaling) • RNA-RNA (regulatory) • Genetic interactions (gene knockouts)

Outline 1. Network Motifs 2. Network integration 3. Network alignment and querying: conserved complexes.

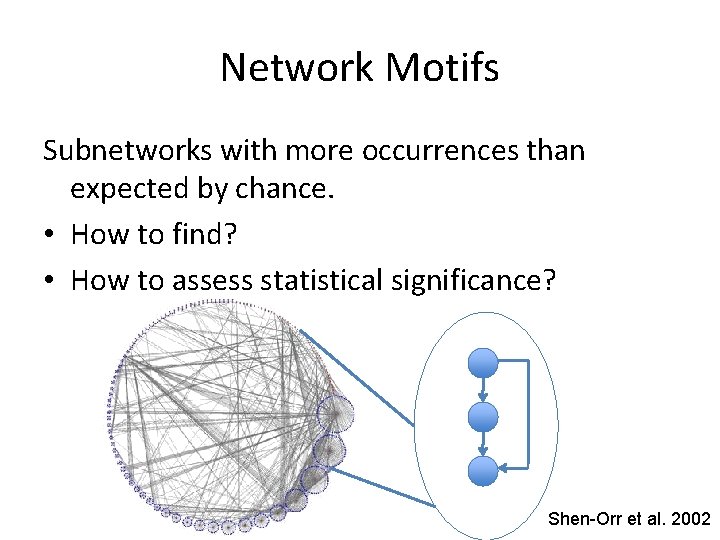
Network Motifs Subnetworks with more occurrences than expected by chance. • How to find? • How to assess statistical significance? Shen-Orr et al. 2002

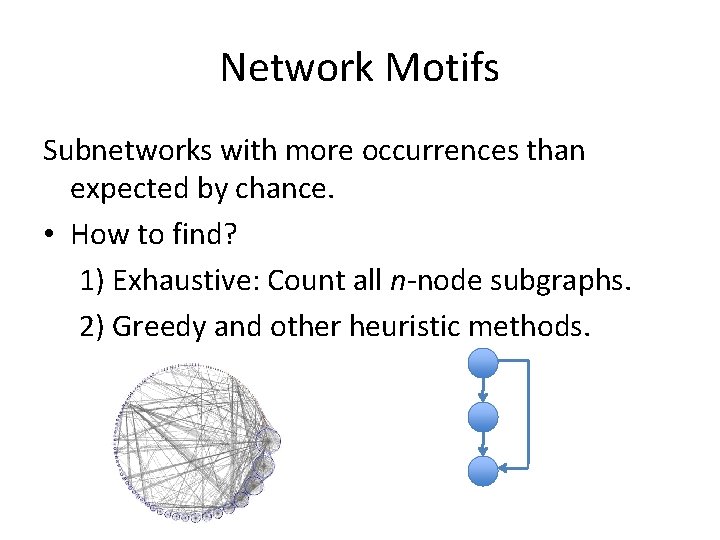
Network Motifs Subnetworks with more occurrences than expected by chance. • How to find? 1) Exhaustive: Count all n-node subgraphs. 2) Greedy and other heuristic methods.

Network Motifs Subnetworks with more occurrences than expected by chance. • How to assess statistical significance? – Compare number of occurrences to random network.

Randomizing a Network Occurrence of motifs depend strongly on network topology. What is an appropriate ensemble of random networks? (null model)

Random Networks Occurrence of motifs depend strongly on network topology. What is an appropriate ensemble of random networks? (null model)

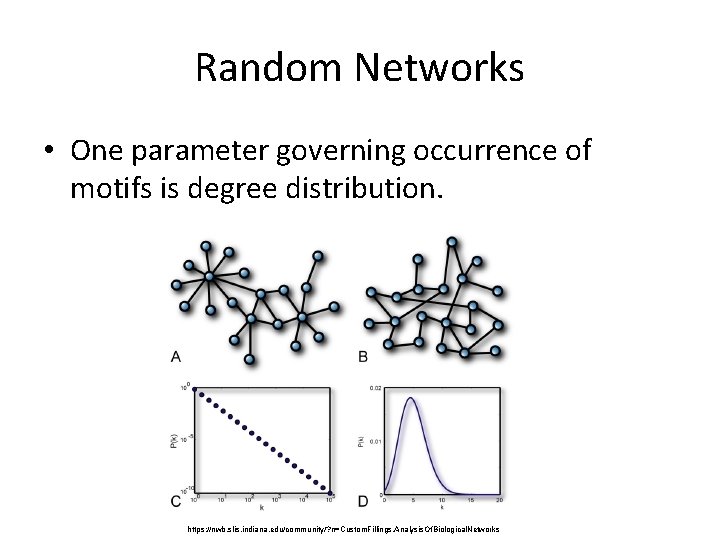
Random Networks • One parameter governing occurrence of motifs is degree distribution. https: //nwb. slis. indiana. edu/community/? n=Custom. Fillings. Analysis. Of. Biological. Networks

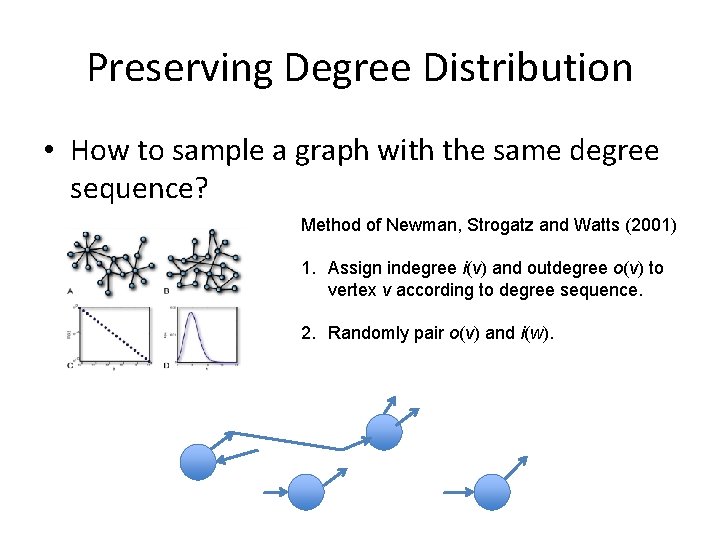
Preserving Degree Distribution • How to sample a graph with the same degree sequence? Method of Newman, Strogatz and Watts (2001) 1. Assign indegree i(v) and outdegree o(v) to vertex v according to degree sequence. 2. Randomly pair o(v) and i(w).

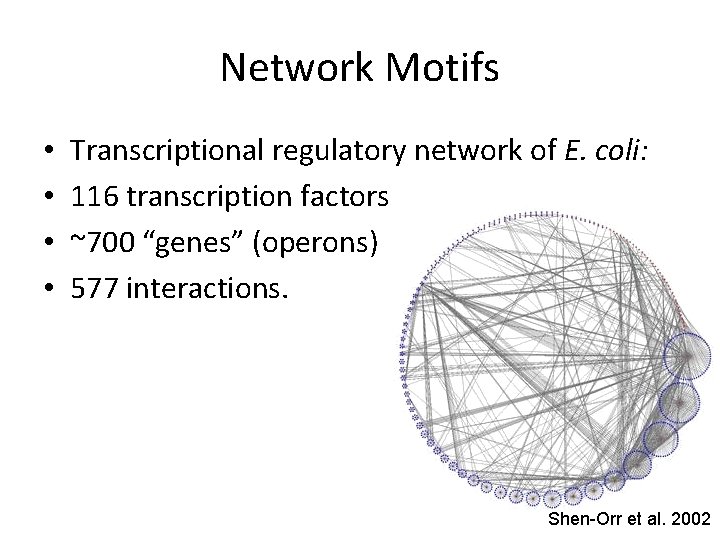
Network Motifs • • Transcriptional regulatory network of E. coli: 116 transcription factors ~700 “genes” (operons) 577 interactions. Shen-Orr et al. 2002

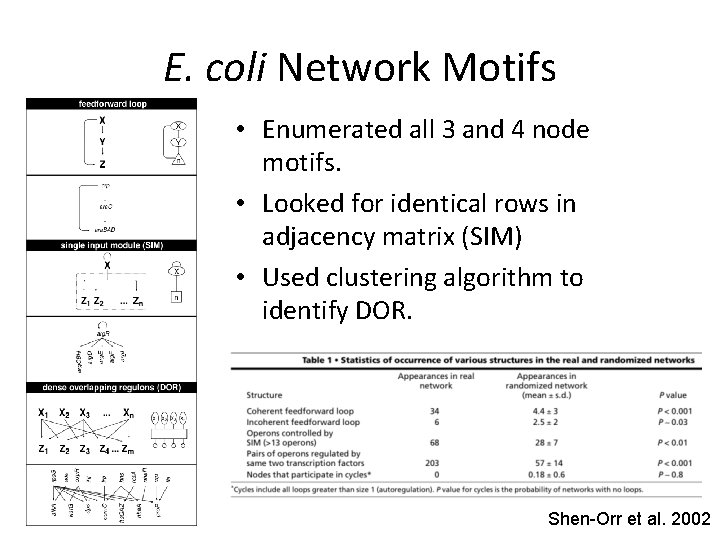
E. coli Network Motifs • Enumerated all 3 and 4 node motifs. • Looked for identical rows in adjacency matrix (SIM) • Used clustering algorithm to identify DOR. Shen-Orr et al. 2002

Importance of Network Motifs • Building block of networks. • Indicate modular structure of biological networks. • Appearance of some motifs might be explained by particular dynamics (e. g. feedforward and feedback loops) • Some skepticism, particularly because data is incomplete.

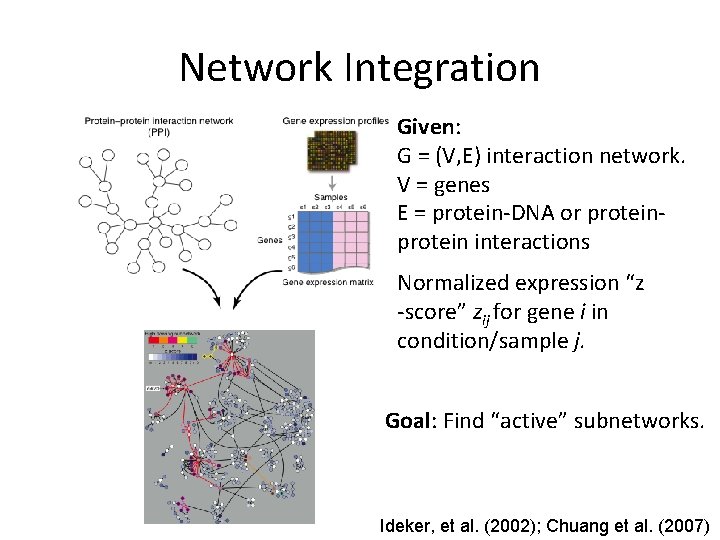
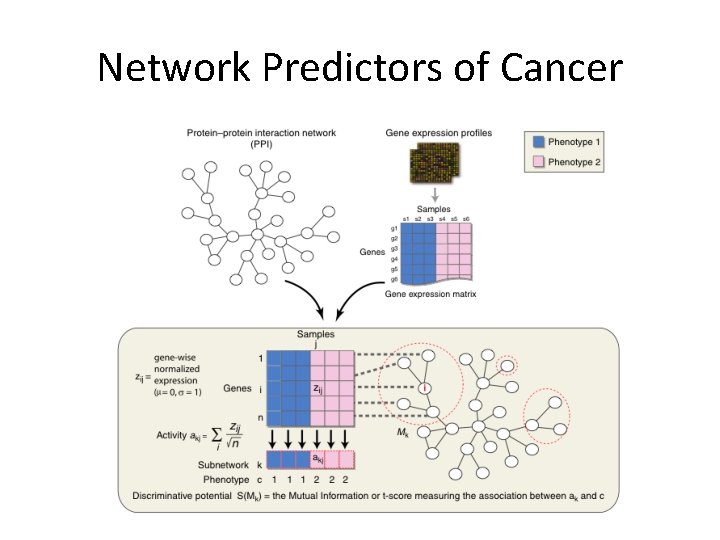
Network Integration Given: G = (V, E) interaction network. V = genes E = protein-DNA or protein interactions Normalized expression “z -score” zij for gene i in condition/sample j. Goal: Find “active” subnetworks. Ideker, et al. (2002); Chuang et al. (2007)

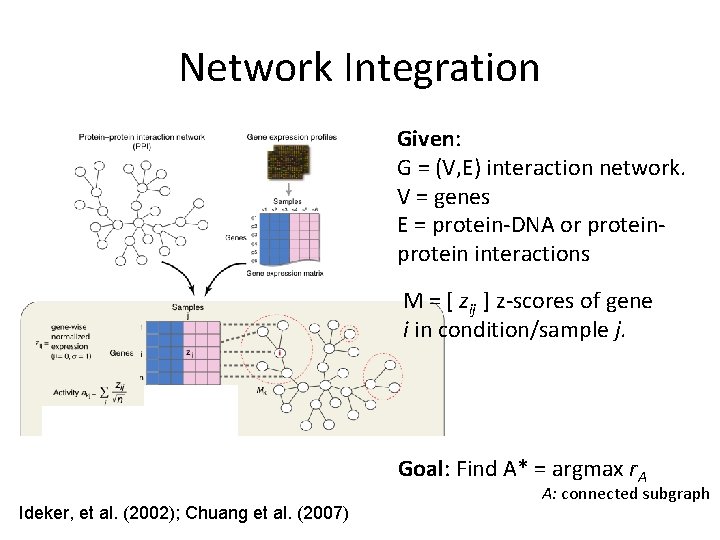
Network Integration Given: G = (V, E) interaction network. V = genes E = protein-DNA or protein interactions M = [ zij ] z-scores of gene i in condition/sample j. Goal: Find A* = argmax r. A Ideker, et al. (2002); Chuang et al. (2007) A: connected subgraph

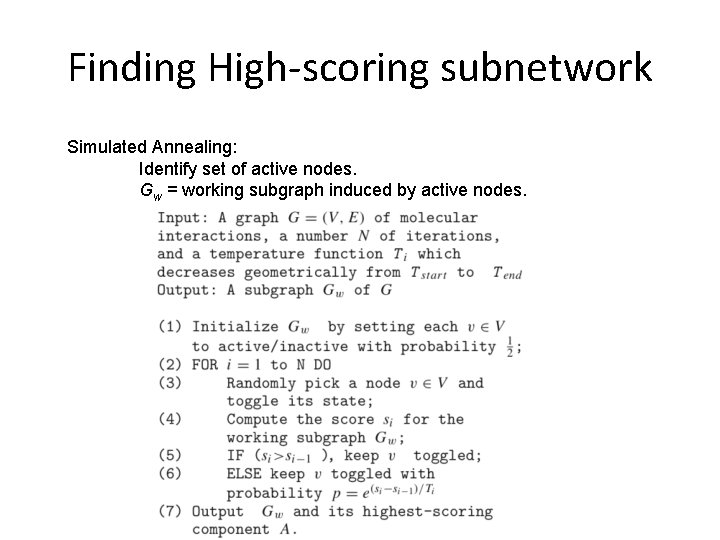
Finding High-scoring subnetwork Simulated Annealing: Identify set of active nodes. Gw = working subgraph induced by active nodes.

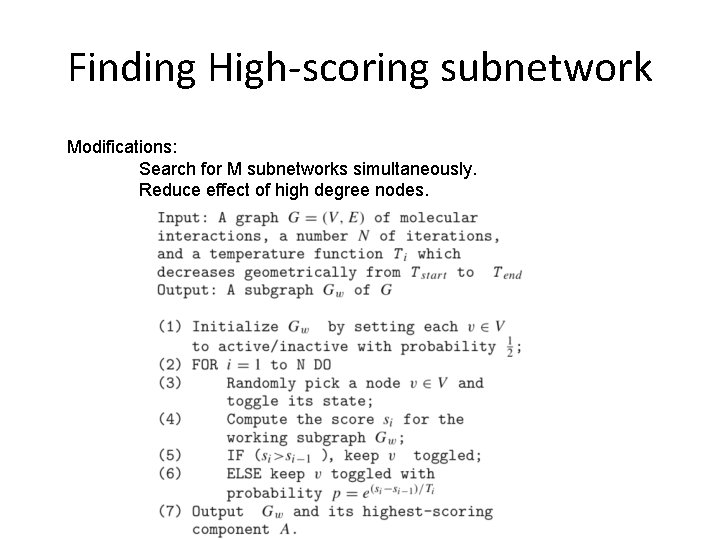
Finding High-scoring subnetwork Modifications: Search for M subnetworks simultaneously. Reduce effect of high degree nodes.

Network Predictors of Cancer

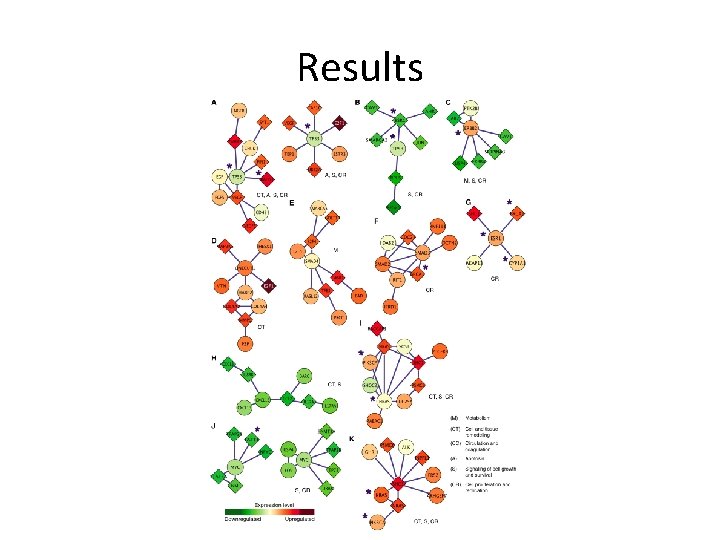
Results

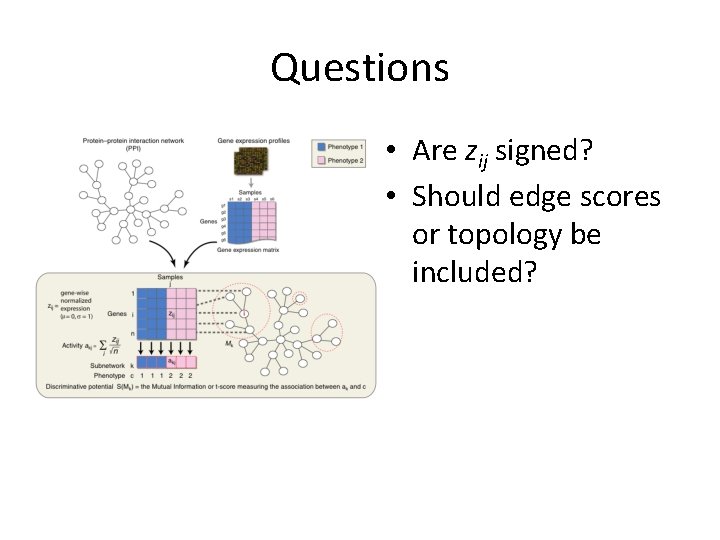
Questions • Are zij signed? • Should edge scores or topology be included?

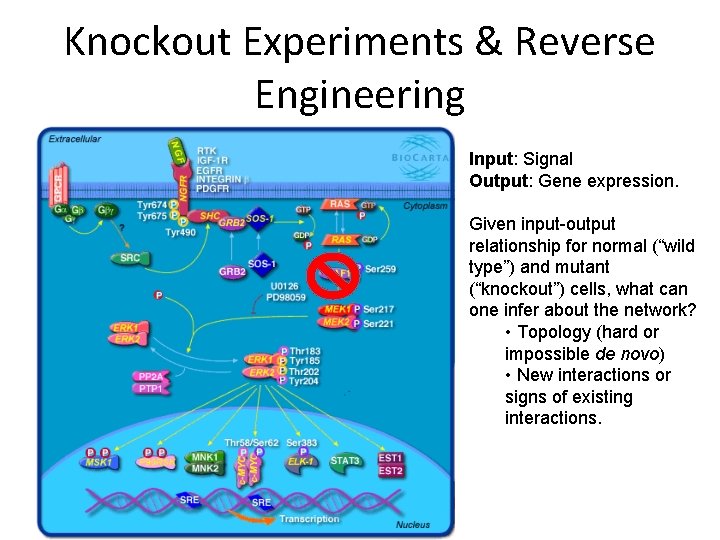
Knockout Experiments & Reverse Engineering Input: Signal Output: Gene expression. Given input-output relationship for normal (“wild type”) and mutant (“knockout”) cells, what can one infer about the network? • Topology (hard or impossible de novo) • New interactions or signs of existing interactions.

Sources • • Shen-Orr, S. S. , Milo, R. , Mangan, S. , et al. 2002. Network motifs in the transcriptional regulation network of Escherichia coli. Nature Genetics 31, 64– 68. Newman, M. E. J. , Strogatz, S. H. , and Watts, D. J. 2001. Random graphs with arbitrary degree distributions and their applications. Phys. Rev. E 64, 026118– 026134. Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002; 18 Suppl 1: S 23340. Chuang HY, Lee E, Liu YT, Lee D, Ideker T. 2007. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007; 3: 140.