Cholesky decomposition May 27 th 2015 Helsinki Finland

![Number of parameters Bivariate [nvar X (nvar+1)] / 2 = number of A paths Number of parameters Bivariate [nvar X (nvar+1)] / 2 = number of A paths](https://slidetodoc.com/presentation_image_h/3175bb590cec7105ab0e47055f5f1374/image-14.jpg)
![Number of parameters Trivariate [nvar X (nvar+1)] / 2 = number of A paths Number of parameters Trivariate [nvar X (nvar+1)] / 2 = number of A paths](https://slidetodoc.com/presentation_image_h/3175bb590cec7105ab0e47055f5f1374/image-23.jpg)




- Slides: 28

Cholesky decomposition May 27 th 2015 Helsinki, Finland E. Vuoksimaa

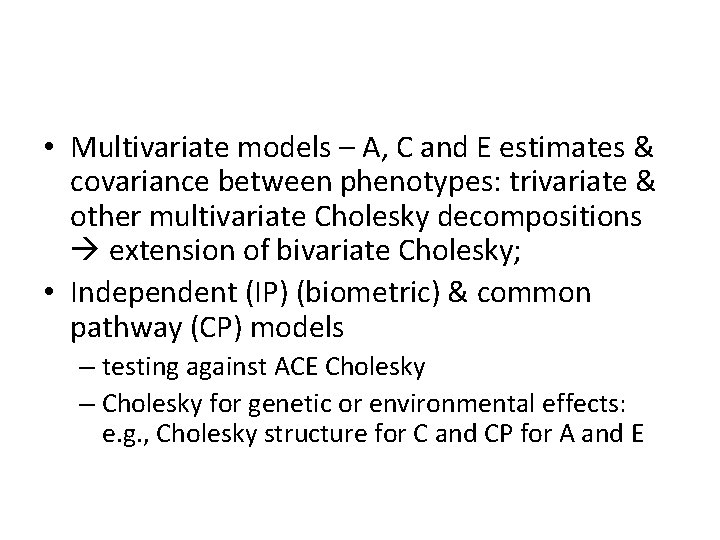
Univariate & multivariate approach • Univariate models – A, C and E estimates • Bivariate Cholesky – A, C and E estimates & covariance between two phenotypes – ACE more power compared to univariate scenario – two interpretations on the relationship between two phenotypes 1) how much of the variance is explained by A, C, E effect that are shared between phenotypes 2) decomposing phenotypic correlation into genetic and environmental correlations

• Multivariate models – A, C and E estimates & covariance between phenotypes: trivariate & other multivariate Cholesky decompositions extension of bivariate Cholesky; • Independent (IP) (biometric) & common pathway (CP) models – testing against ACE Cholesky – Cholesky for genetic or environmental effects: e. g. , Cholesky structure for C and CP for A and E

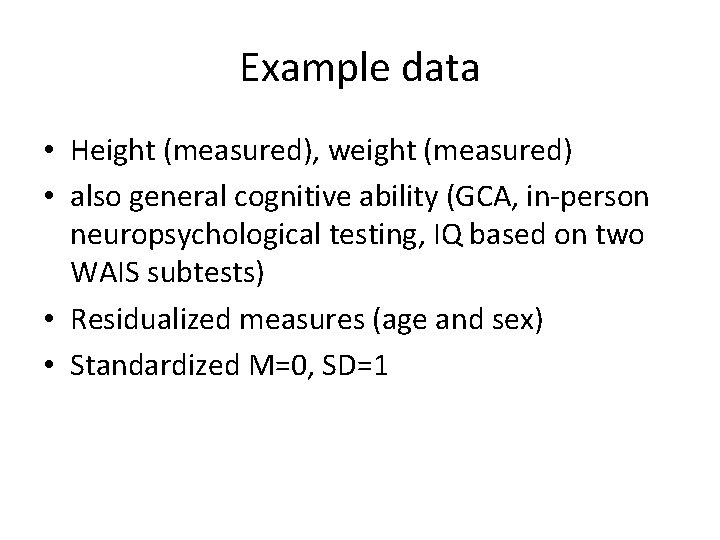
Example data • Height (measured), weight (measured) • also general cognitive ability (GCA, in-person neuropsychological testing, IQ based on two WAIS subtests) • Residualized measures (age and sex) • Standardized M=0, SD=1

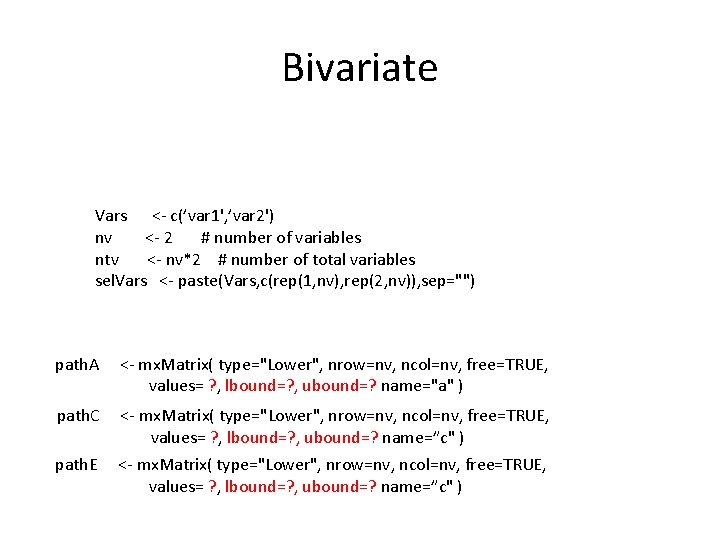
Bivariate Vars <- c(’var 1', ’var 2') nv <- 2 # number of variables ntv <- nv*2 # number of total variables sel. Vars <- paste(Vars, c(rep(1, nv), rep(2, nv)), sep="") path. A <- mx. Matrix( type="Lower", nrow=nv, ncol=nv, free=TRUE, values= ? , lbound=? , ubound=? name="a" ) path. C <- mx. Matrix( type="Lower", nrow=nv, ncol=nv, free=TRUE, values= ? , lbound=? , ubound=? name=”c" ) path. E <- mx. Matrix( type="Lower", nrow=nv, ncol=nv, free=TRUE, values= ? , lbound=? , ubound=? name=”c" )

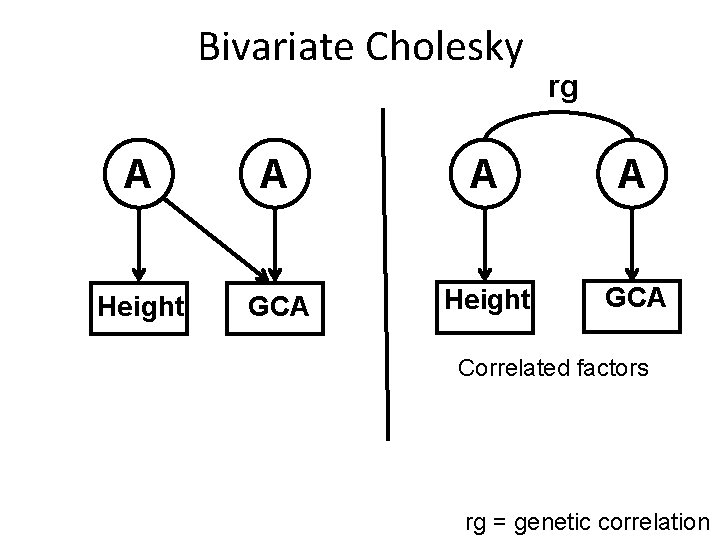
Bivariate Cholesky rg A A Height GCA Correlated factors rg = genetic correlation

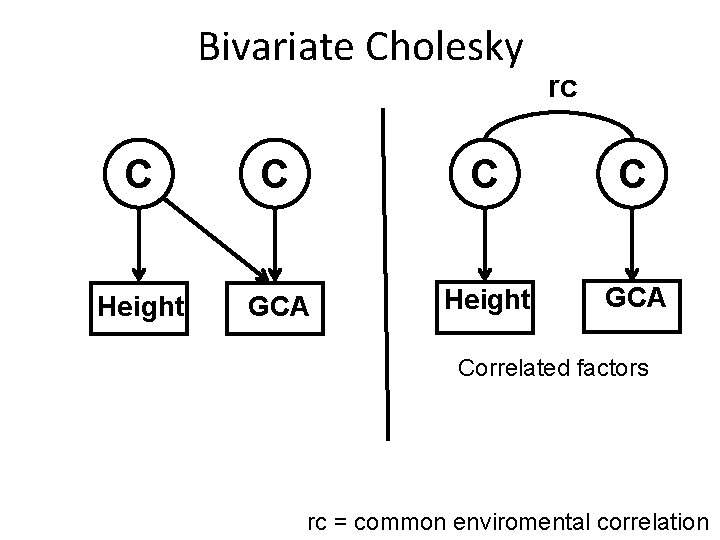
Bivariate Cholesky rc C C Height GCA Correlated factors rc = common enviromental correlation

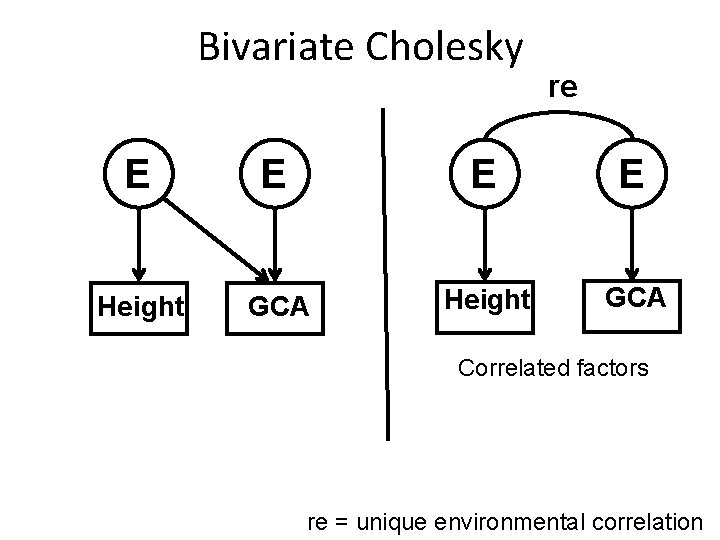
Bivariate Cholesky re E E Height GCA Correlated factors re = unique environmental correlation

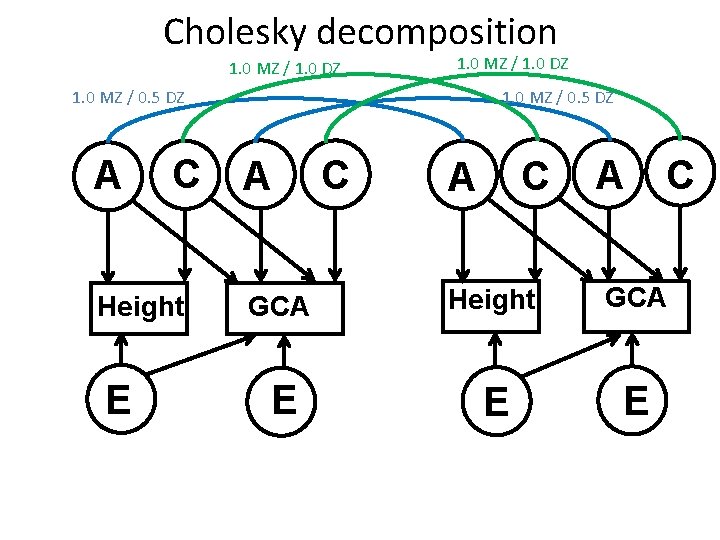
Cholesky decomposition 1. 0 MZ / 1. 0 DZ 1. 0 MZ / 0. 5 DZ A C Height E 1. 0 MZ / 0. 5 DZ A C C A A C GCA Height GCA E E E

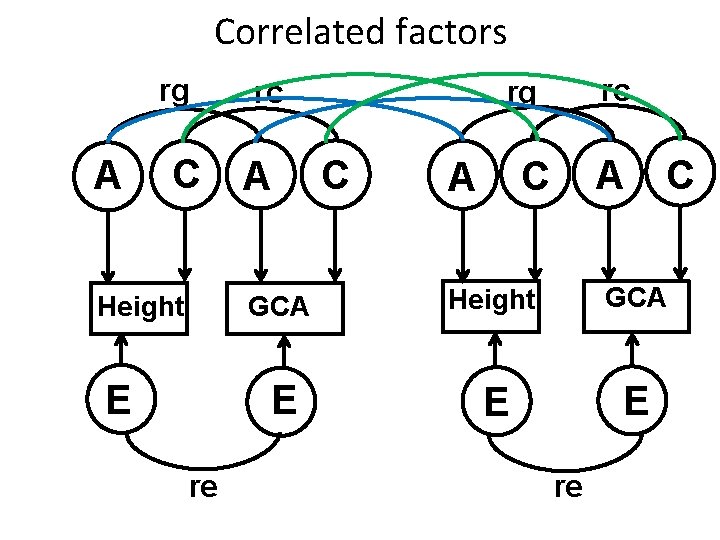
Correlated factors rg A C Height E re A rc rg rc C A C GCA Height GCA E E E re

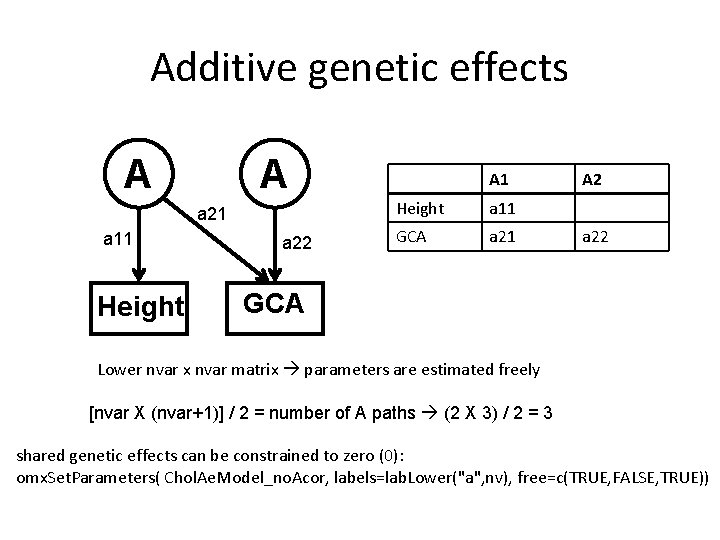
Additive genetic effects A A a 21 a 11 Height a 22 A 1 Height a 11 GCA a 21 A 2 a 22 GCA Lower nvar x nvar matrix parameters are estimated freely [nvar X (nvar+1)] / 2 = number of A paths (2 X 3) / 2 = 3 shared genetic effects can be constrained to zero (0): omx. Set. Parameters( Chol. Ae. Model_no. Acor, labels=lab. Lower("a", nv), free=c(TRUE, FALSE, TRUE))

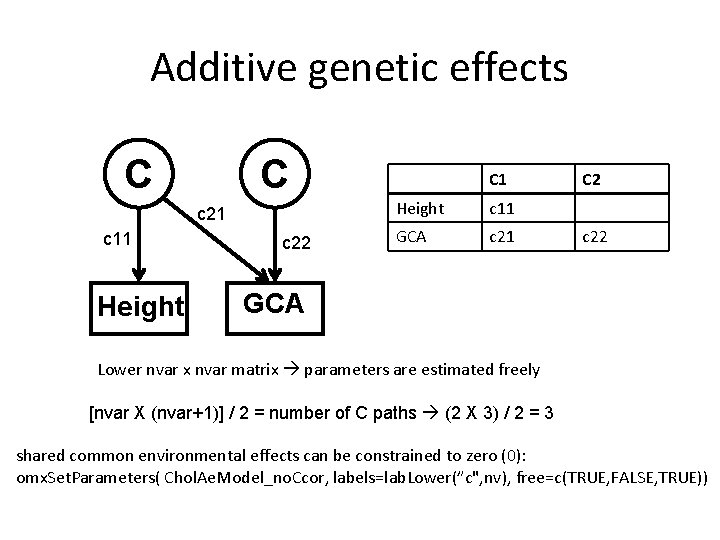
Additive genetic effects C C c 21 c 11 Height c 22 C 1 Height c 11 GCA c 21 C 2 c 22 GCA Lower nvar x nvar matrix parameters are estimated freely [nvar X (nvar+1)] / 2 = number of C paths (2 X 3) / 2 = 3 shared common environmental effects can be constrained to zero (0): omx. Set. Parameters( Chol. Ae. Model_no. Ccor, labels=lab. Lower(”c", nv), free=c(TRUE, FALSE, TRUE))

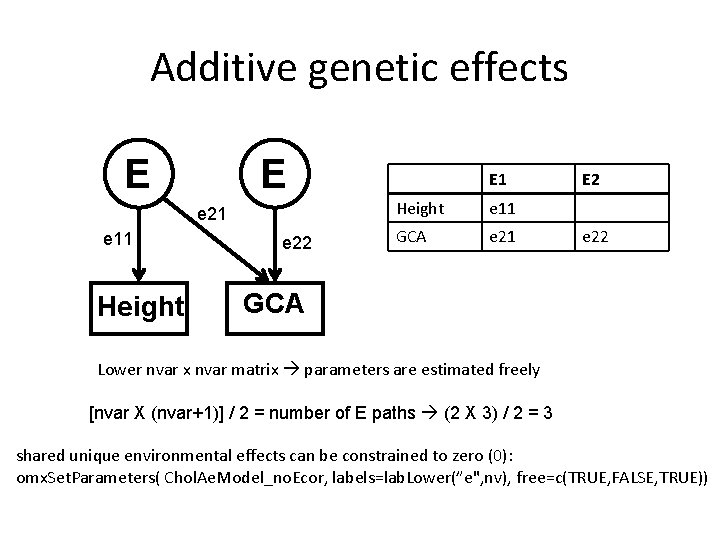
Additive genetic effects E E e 21 e 11 Height e 22 E 1 Height e 11 GCA e 21 E 2 e 22 GCA Lower nvar x nvar matrix parameters are estimated freely [nvar X (nvar+1)] / 2 = number of E paths (2 X 3) / 2 = 3 shared unique environmental effects can be constrained to zero (0): omx. Set. Parameters( Chol. Ae. Model_no. Ecor, labels=lab. Lower(”e", nv), free=c(TRUE, FALSE, TRUE))
![Number of parameters Bivariate nvar X nvar1 2 number of A paths Number of parameters Bivariate [nvar X (nvar+1)] / 2 = number of A paths](https://slidetodoc.com/presentation_image_h/3175bb590cec7105ab0e47055f5f1374/image-14.jpg)
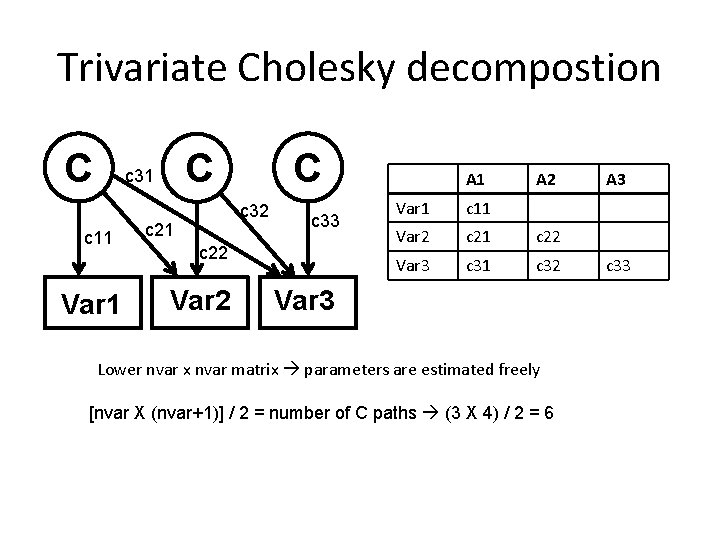
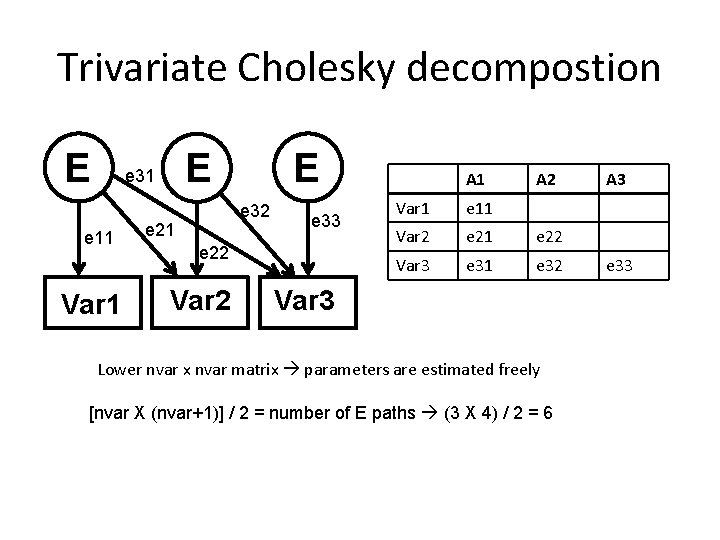
Number of parameters Bivariate [nvar X (nvar+1)] / 2 = number of A paths (2 X 3) / 2 = 3 [nvar X (nvar+1)] / 2 = number of C paths (2 X 3) / 2 = 3 [nvar X (nvar+1)] / 2 = number of E paths (2 X 3) / 2 = 3 means = 2 number of parameters = 11 Number of parameters in AE-AE cholesky ? Number of parameters in AE-AE cholesky where rg = 0 ?

Proportion of phenotypic correlation due to rg • (√a 2 var 1 X rg X √a 2 var 2) / rp • (√ heritability of phenotype 1 X genetic correlation between phenotype 1 and phenotype 2 X √ heritability of phenotype 2) / phenotypic correlation

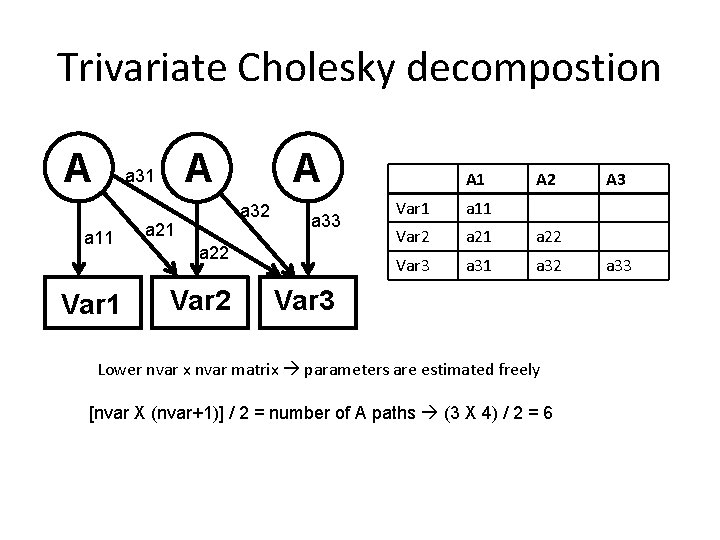
Trivariate Vars <- c(’var 1', ’var 2’, ’var 3’) # add 3 rd variable, 4 th, 5 th, etc. nv <- 3 # number of variables # you need to change this, here 3 ntv <- nv*2 # number of total variables sel. Vars <- paste(Vars, c(rep(1, nv), rep(2, nv)), sep="") Matrices to get the path coefficients for a, c and e path. A <- mx. Matrix( type="Lower", nrow=nv, ncol=nv, free=TRUE, values= ? , lbound=? , ubound=? name="a" ) path. C <- mx. Matrix( type="Lower", nrow=nv, ncol=nv, free=TRUE, values= ? , lbound=? , ubound=? name=”c" ) path. E <- mx. Matrix( type="Lower", nrow=nv, ncol=nv, free=TRUE, values= ? , lbound=? , ubound=? name=”c" )

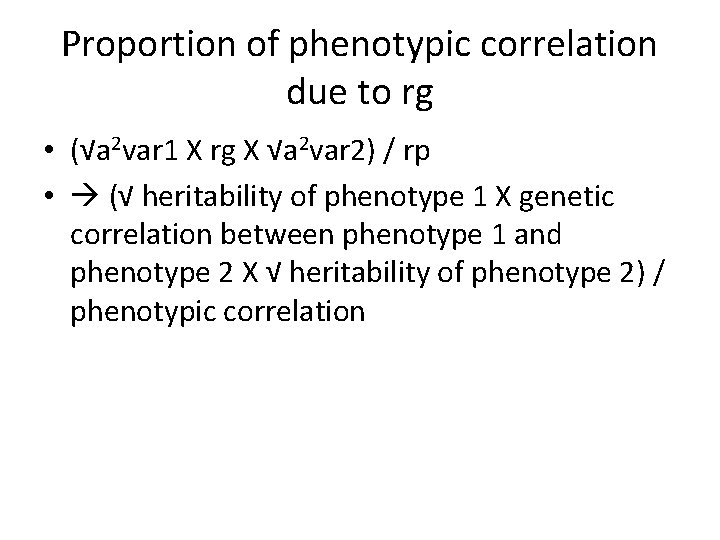
Trivariate Cholesky decompostion A a 11 Var 1 A A a 31 a 32 a 21 a 33 a 22 Var 2 A 1 A 2 Var 1 a 11 Var 2 a 21 a 22 Var 3 a 31 a 32 Var 3 Lower nvar x nvar matrix parameters are estimated freely [nvar X (nvar+1)] / 2 = number of A paths (3 X 4) / 2 = 6 A 3 a 33

Trivariate Cholesky decompostion C c 11 Var 1 C C c 31 c 32 c 21 c 33 c 22 Var 2 A 1 A 2 Var 1 c 11 Var 2 c 21 c 22 Var 3 c 31 c 32 Var 3 Lower nvar x nvar matrix parameters are estimated freely [nvar X (nvar+1)] / 2 = number of C paths (3 X 4) / 2 = 6 A 3 c 33

Trivariate Cholesky decompostion E e 11 Var 1 E E e 31 e 32 e 21 e 33 e 22 Var 2 A 1 A 2 Var 1 e 11 Var 2 e 21 e 22 Var 3 e 31 e 32 Var 3 Lower nvar x nvar matrix parameters are estimated freely [nvar X (nvar+1)] / 2 = number of E paths (3 X 4) / 2 = 6 A 3 e 33

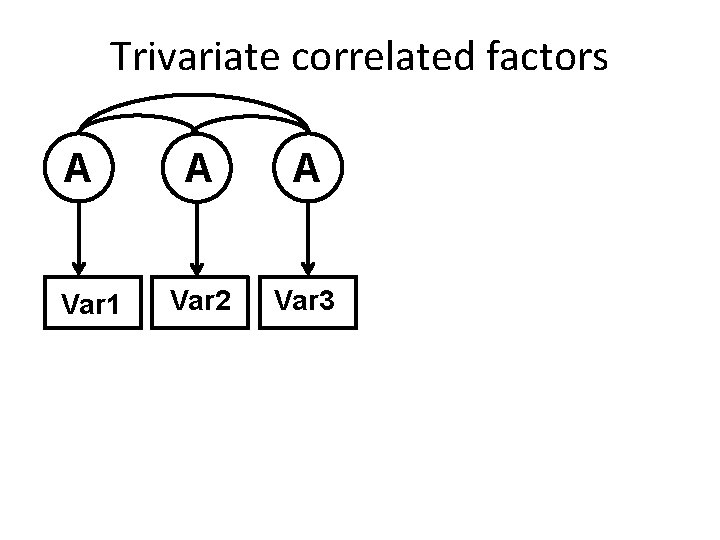
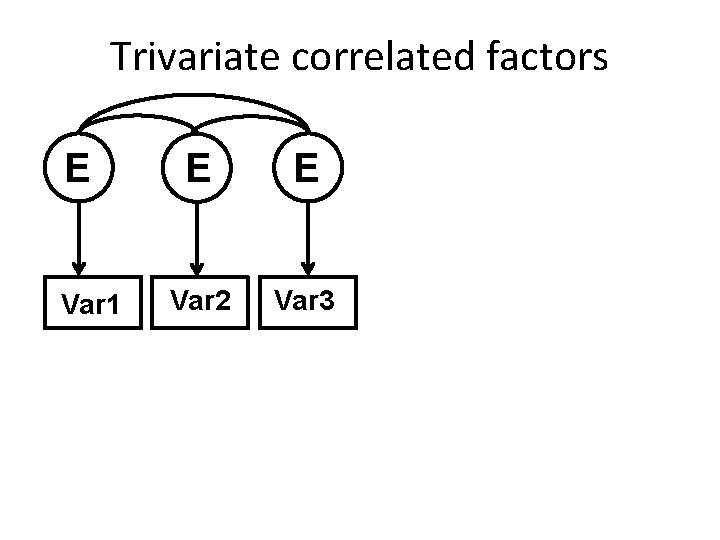
Trivariate correlated factors A Var 1 A A Var 2 Var 3

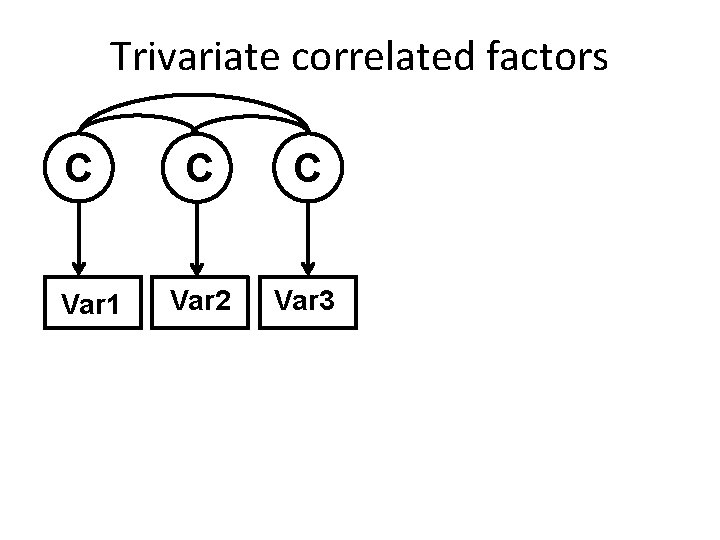
Trivariate correlated factors C Var 1 C C Var 2 Var 3

Trivariate correlated factors E Var 1 E E Var 2 Var 3
![Number of parameters Trivariate nvar X nvar1 2 number of A paths Number of parameters Trivariate [nvar X (nvar+1)] / 2 = number of A paths](https://slidetodoc.com/presentation_image_h/3175bb590cec7105ab0e47055f5f1374/image-23.jpg)
Number of parameters Trivariate [nvar X (nvar+1)] / 2 = number of A paths [nvar X (nvar+1)] / 2 = number of C paths [nvar X (nvar+1)] / 2 = number of E paths means number of parameters = ?

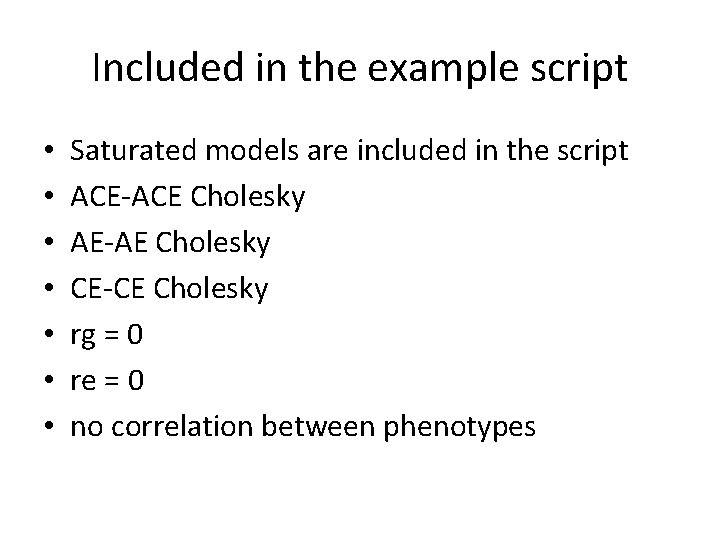
Included in the example script • • Saturated models are included in the script ACE-ACE Cholesky AE-AE Cholesky CE-CE Cholesky rg = 0 re = 0 no correlation between phenotypes

• Bivariate with height & weight, also height & GCA and weight & GCA • Calculate genetic and environmental correlations • Can we set rg/re or both as zero? • What is the proportion of phenotypic correlation due to rg?

Things to consider • Do not automatically run AE-AE after ACE-ACE, e. g. , consider if you want to keep C effects for one(/some) of the variables • E. g. , C effects of about 15% may be fixed to be zero, but you may still want to keep the C effects – less biased genetic correlation • Cholesky in context of IP and CP models • What is the question that you are asking

Suggested reading • Carey G. (1988), Behavior Genetics, 18, 329 -338. • Loehlin (1996). The Cholesky approach: a cautionary note. Behavior Genetics, 26, 65 -69. • Carey G. (2005). Cholesky problems. Behavior Genetics, 35, 653 -665. • Wu and Neale (2013). On the likelihood ratio tests in bivariate ACDE models. Psychometrika, 78, 441 -463. • Panizzon et al. (2014). Genetic and environmental influences on general cognitive ability: is g a valid latent construct. Intelligence, 43, 65 -76.

Resources including presentations • International Twin workshop, every March, Institute for behavioral genetics, University of Boulder Colorado • QIMR, Workshop, Sarah Medland
 Cholesky decomposition excel
Cholesky decomposition excel Interflora finland helsinki
Interflora finland helsinki Helena egan
Helena egan Helsinki business polytechnic
Helsinki business polytechnic Adoption in finland
Adoption in finland Kvanttielektrodynamiikka
Kvanttielektrodynamiikka Finland quality design
Finland quality design Tratado de helsinque
Tratado de helsinque Biocenter finland
Biocenter finland Yksityinen vs julkinen terveydenhuolto
Yksityinen vs julkinen terveydenhuolto Institute for the languages of finland
Institute for the languages of finland What is the origin of the word easter
What is the origin of the word easter Micro research finland
Micro research finland Aikuiskasvatustiede helsinki
Aikuiskasvatustiede helsinki Pisa
Pisa Uusimaa avi
Uusimaa avi Tupas finland
Tupas finland Kieleily
Kieleily Testbed helsinki
Testbed helsinki Inner wheel finland
Inner wheel finland Tarramitta
Tarramitta Finnish national board of education
Finnish national board of education Helsinki university physics
Helsinki university physics Helsinki lappeenranta
Helsinki lappeenranta Finlanda capitala
Finlanda capitala Kemh13
Kemh13 Dressmann finland
Dressmann finland Ngonorway
Ngonorway Lennot helsinki antananarivo
Lennot helsinki antananarivo