Ch IPSeq Data Processing and QC Simon Andrews


- Slides: 17

Ch. IP-Seq Data Processing and QC Simon Andrews simon. andrews@babraham. ac. uk @simon_andrews v 2020 -05

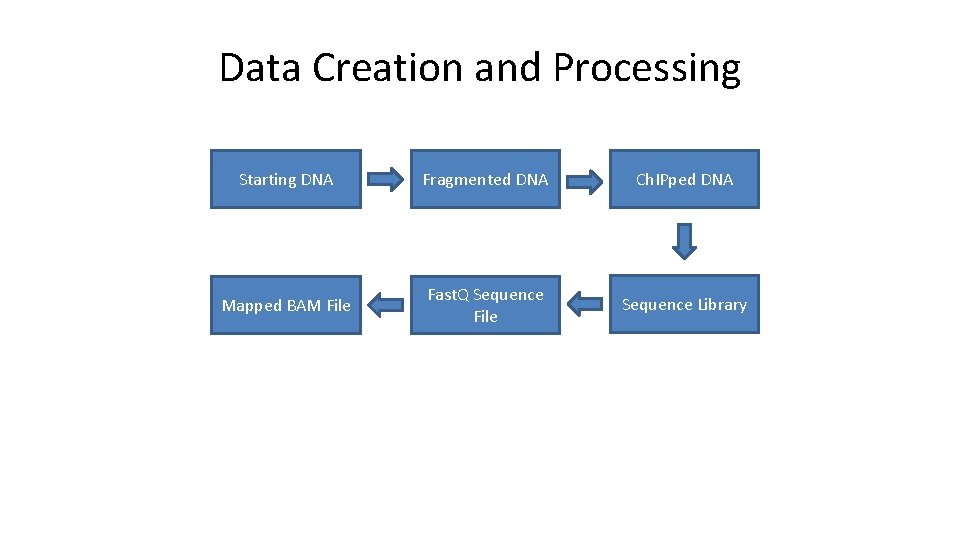
Data Creation and Processing Starting DNA Fragmented DNA Ch. IPped DNA Mapped BAM File Fast. Q Sequence File Sequence Library

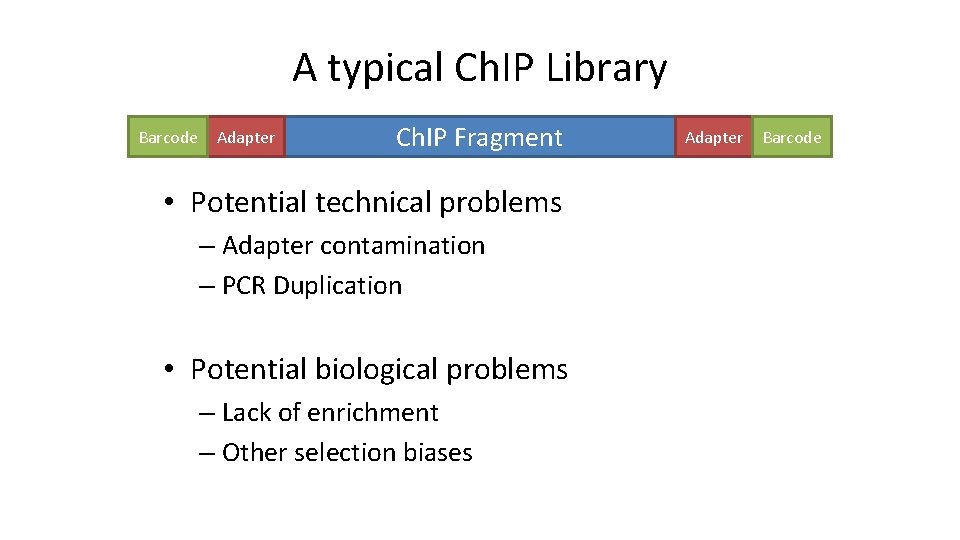
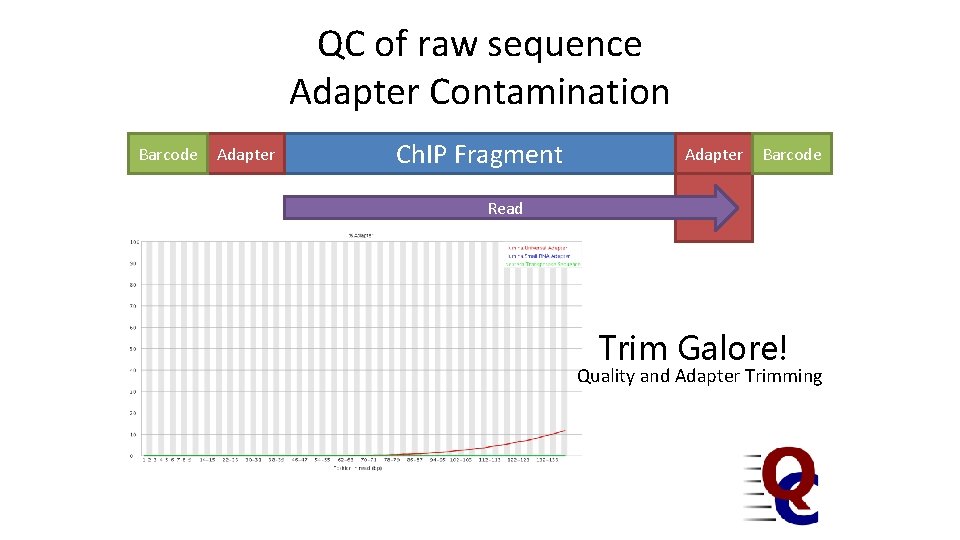
A typical Ch. IP Library Barcode Adapter Ch. IP Fragment • Potential technical problems – Adapter contamination – PCR Duplication • Potential biological problems – Lack of enrichment – Other selection biases Adapter Barcode

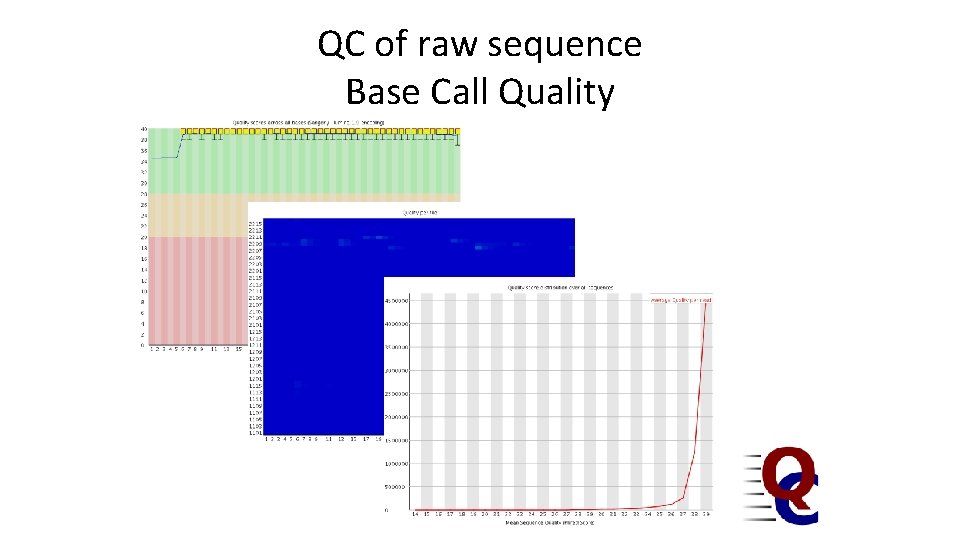
QC of raw sequence Base Call Quality

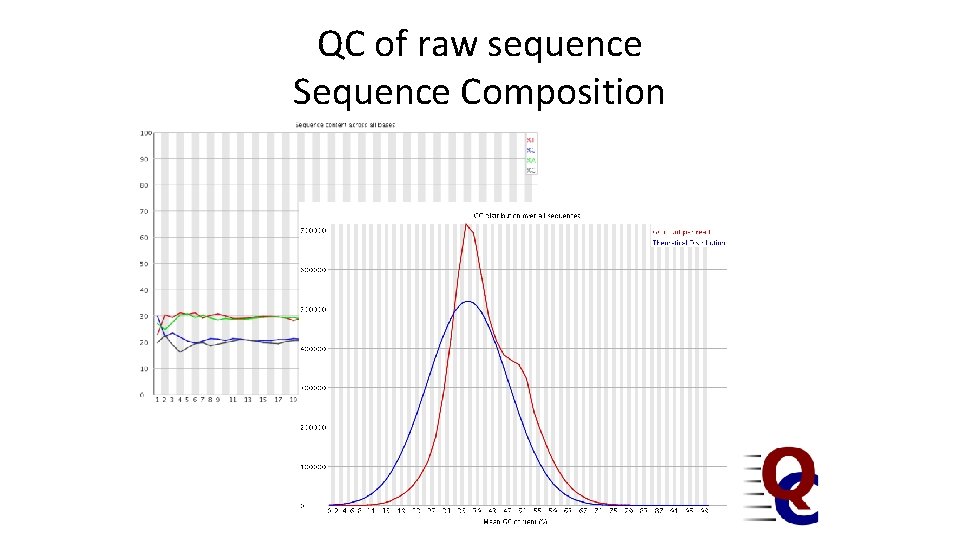
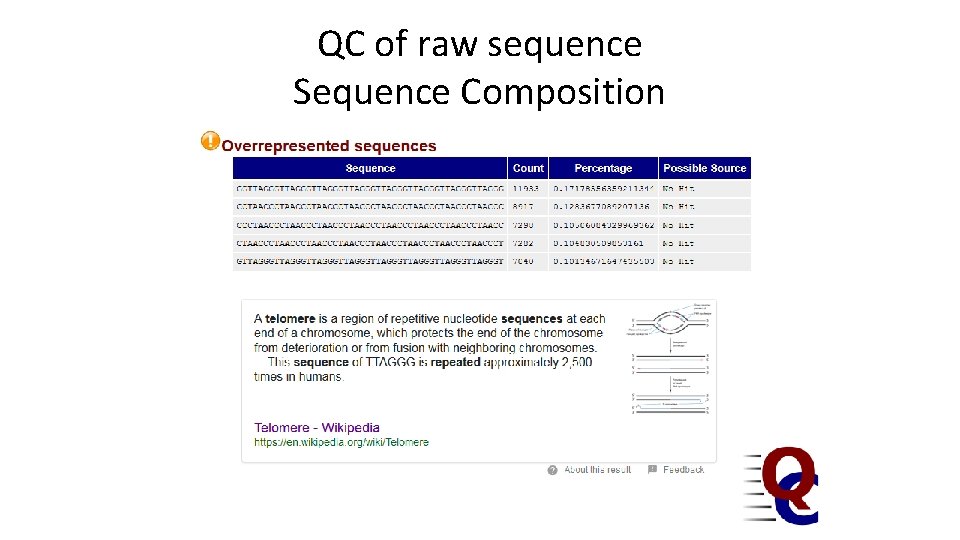
QC of raw sequence Sequence Composition

QC of raw sequence Sequence Composition

QC of raw sequence Adapter Contamination Barcode Adapter Ch. IP Fragment Adapter Barcode Read Trim Galore! Quality and Adapter Trimming

Mapping Ch. IP Data • All regions should be linear genomic stretches • Standard genomic aligners are fine – Bowtie 2 – BWA http: //bowtie-bio. sourceforge. net/bowtie 2/ http: //bio-bwa. sourceforge. net/

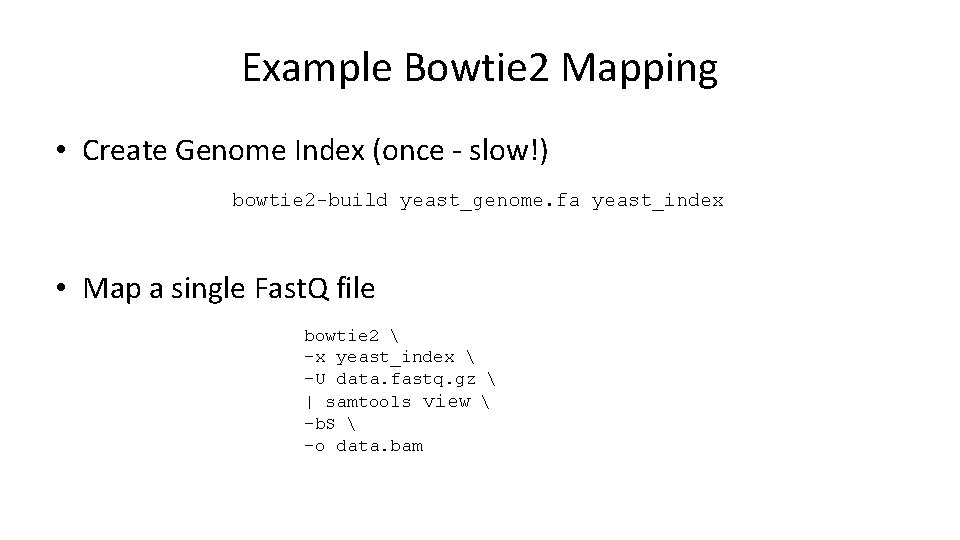
Example Bowtie 2 Mapping • Create Genome Index (once - slow!) bowtie 2 -build yeast_genome. fa yeast_index • Map a single Fast. Q file bowtie 2 -x yeast_index -U data. fastq. gz | samtools view -b. S -o data. bam

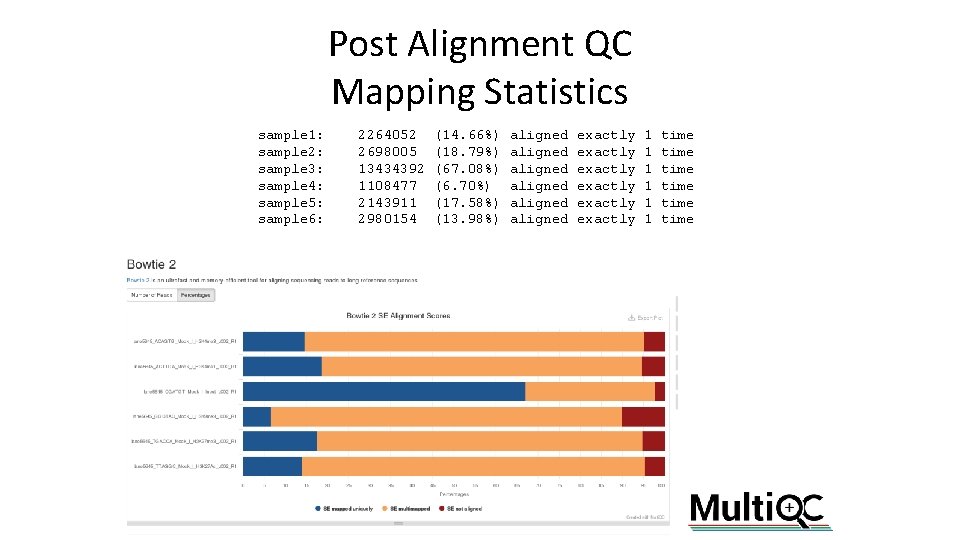
Post Alignment QC Mapping Statistics sample 1: sample 2: sample 3: sample 4: sample 5: sample 6: 2264052 2698005 13434392 1108477 2143911 2980154 (14. 66%) (18. 79%) (67. 08%) (6. 70%) (17. 58%) (13. 98%) aligned aligned exactly exactly 1 1 1 time time

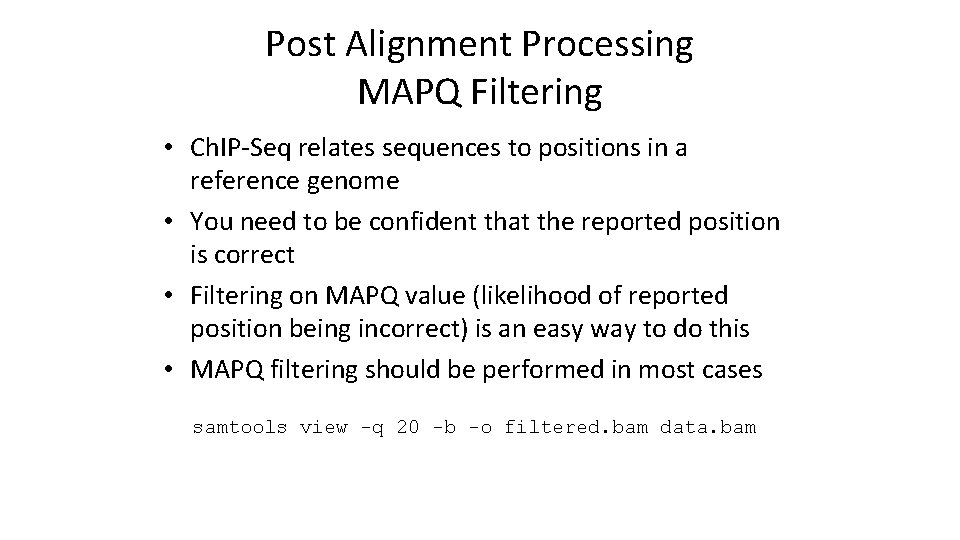
Post Alignment Processing MAPQ Filtering • Ch. IP-Seq relates sequences to positions in a reference genome • You need to be confident that the reported position is correct • Filtering on MAPQ value (likelihood of reported position being incorrect) is an easy way to do this • MAPQ filtering should be performed in most cases samtools view -q 20 -b -o filtered. bam data. bam

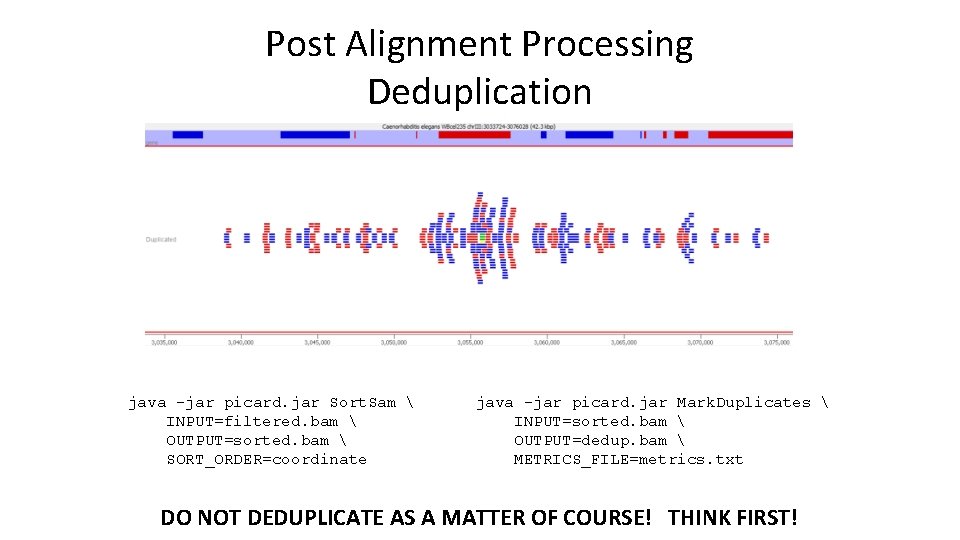
Post Alignment Processing Deduplication java -jar picard. jar Sort. Sam INPUT=filtered. bam OUTPUT=sorted. bam SORT_ORDER=coordinate java -jar picard. jar Mark. Duplicates INPUT=sorted. bam OUTPUT=dedup. bam METRICS_FILE=metrics. txt DO NOT DEDUPLICATE AS A MATTER OF COURSE! THINK FIRST!

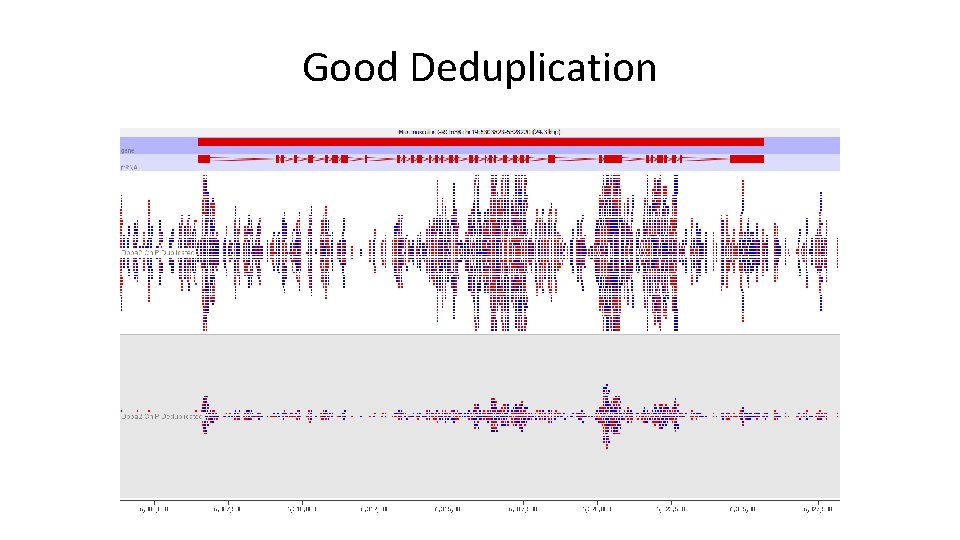
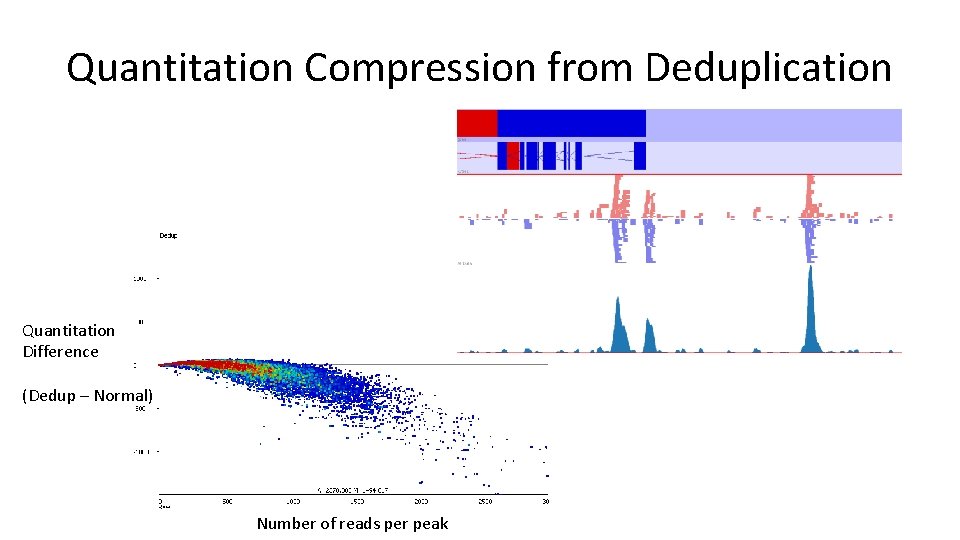
To Deduplicate or Not? • Deduplication can make enrichment visually clearer and help to spot truly enriched regions • Why not just deduplicate everything? – Quantitation compression

Good Deduplication

Quantitation Compression from Deduplication Quantitation Difference (Dedup – Normal) Number of reads per peak

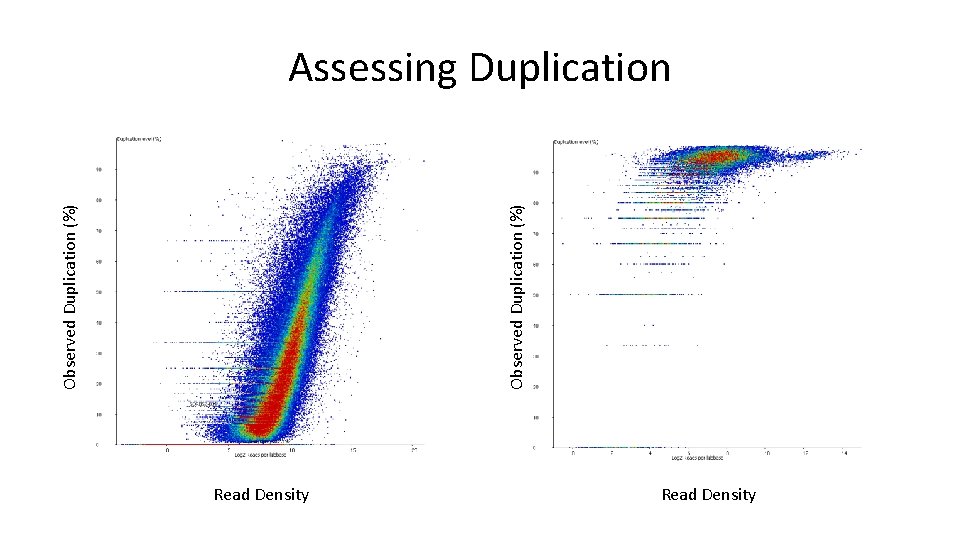
Observed Duplication (%) Assessing Duplication Read Density

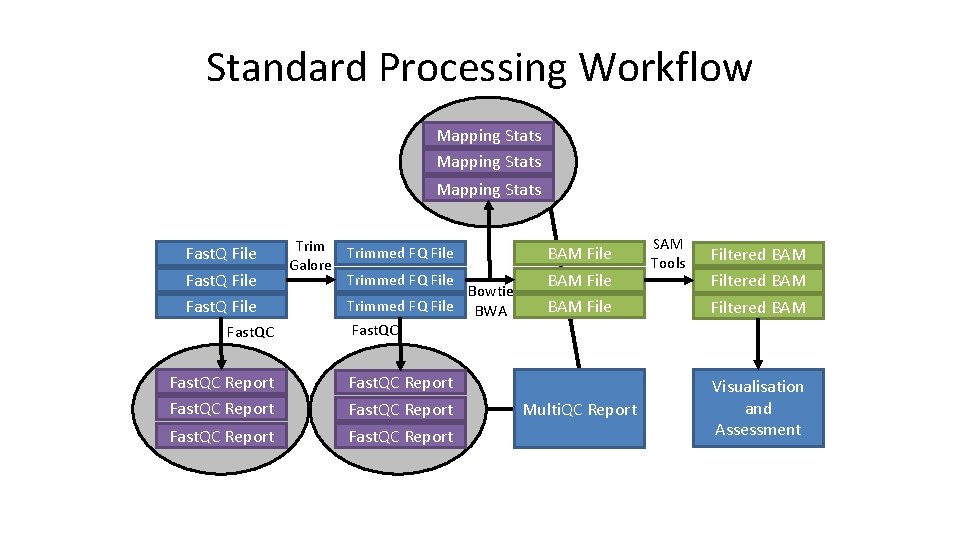
Standard Processing Workflow Mapping Stats Fast. Q File Fast. QC Report Trimmed FQ File Galore Trimmed FQ File Bowtie BWA BAM File SAM Tools Filtered BAM Fast. QC Report Multi. QC Report Visualisation and Assessment