Ch IPSeq Analysis Using CLCGenomics Workbench NOV 16

Ch. IP-Seq Analysis – Using CLCGenomics Workbench NOV 16, 2 017 ANSUM AN CHA TT OPADHYAY, PHD HEALTH SCIE NCE S LIB RA RY SYSTEM UNIVE RS ITY OF PITT SB URGH ANSU MAN@ PITT. EDU

Topics • Transcription Factor Ch. IP-Seq • Histone Ch. IP-Seq • ATAC-Seq

www. hsls. libguides. com/chipseq

Transcription Factor and Histone Ch. IP-Seq
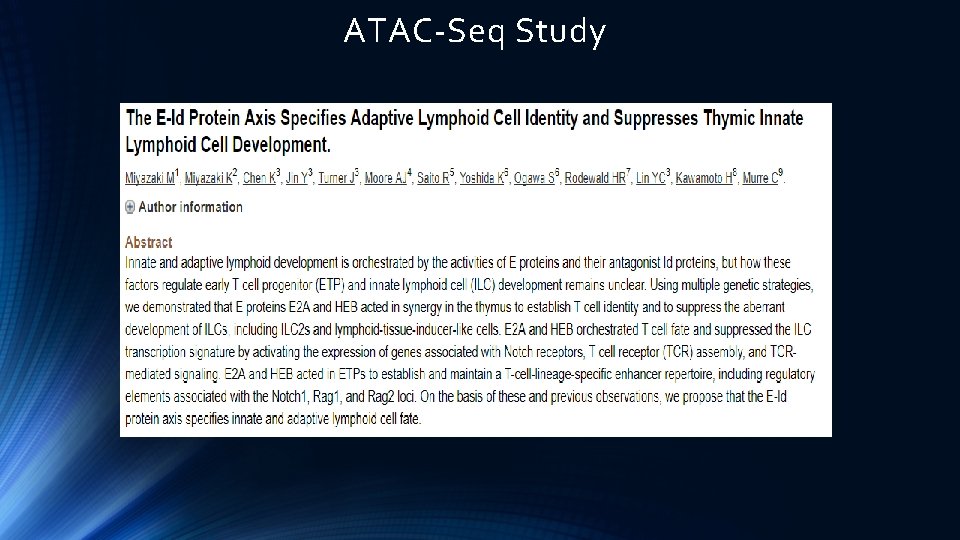
ATAC-Seq Study
Graphical User Interface based software Galaxy : http: //galaxy. crc. pitt. edu: 8080/ CLC Genomics Workbench
Software @ HSLS Mol. Bio http: //hsls. libguides. com/molbio/licensedtools/resources
NGS Software @ HSLS Mol. Bio NGS Analysis Sanger Seq Analysis Human , Mouse and Rat NGS Analysis

CLCbio Genomics Workbench System Requirements • Windows Vista, Windows 7, Windows 8, Windows 10, Windows Server 2008, or Windows Server 2012 • Mac OS X 10. 7 or later. • Linux: Red Hat 5. 0 or later. SUSE 10. 2 or later. Fedora 6 or later. • 8 GB RAM required • 16 GB RAM recommended • 1024 x 768 display required • 1600 x 1200 display recommended • Intel or AMD CPU required • Minimum 10 GB free disc space in the tmp directory
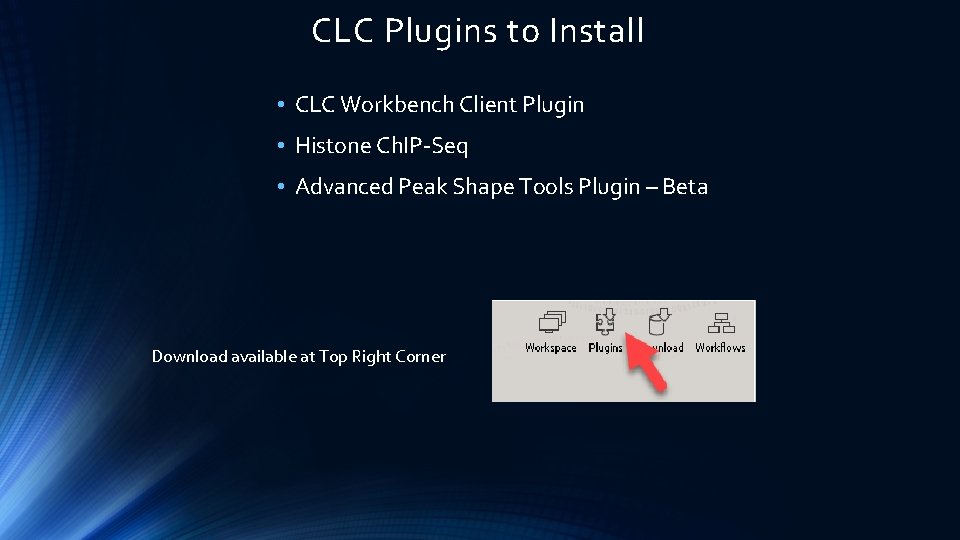
CLC Plugins to Install • CLC Workbench Client Plugin • Histone Ch. IP-Seq • Advanced Peak Shape Tools Plugin – Beta Download available at Top Right Corner
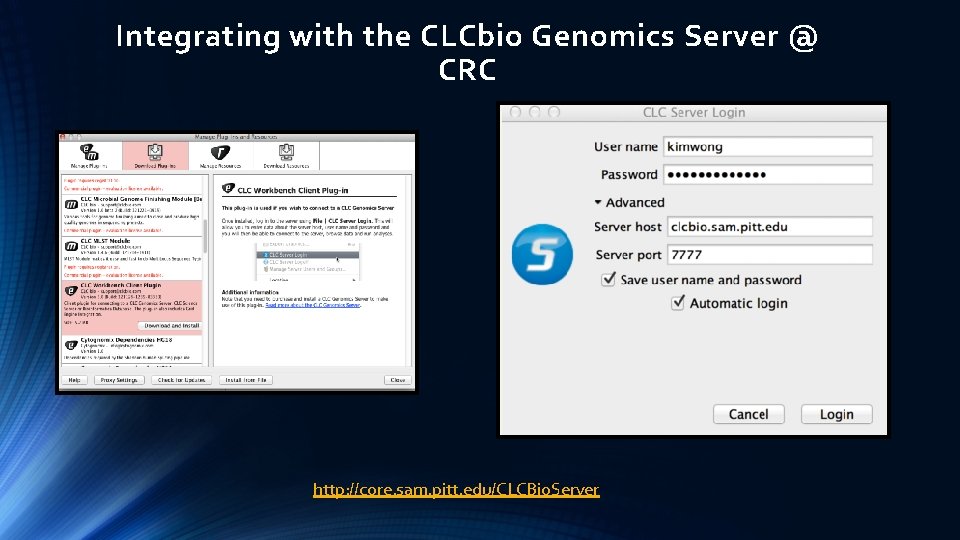
Integrating with the CLCbio Genomics Server @ CRC http: //core. sam. pitt. edu/CLCBio. Server
You need Secure Remote Access via Pulse to run CLCGx from off campus locations / Pitt Wireless
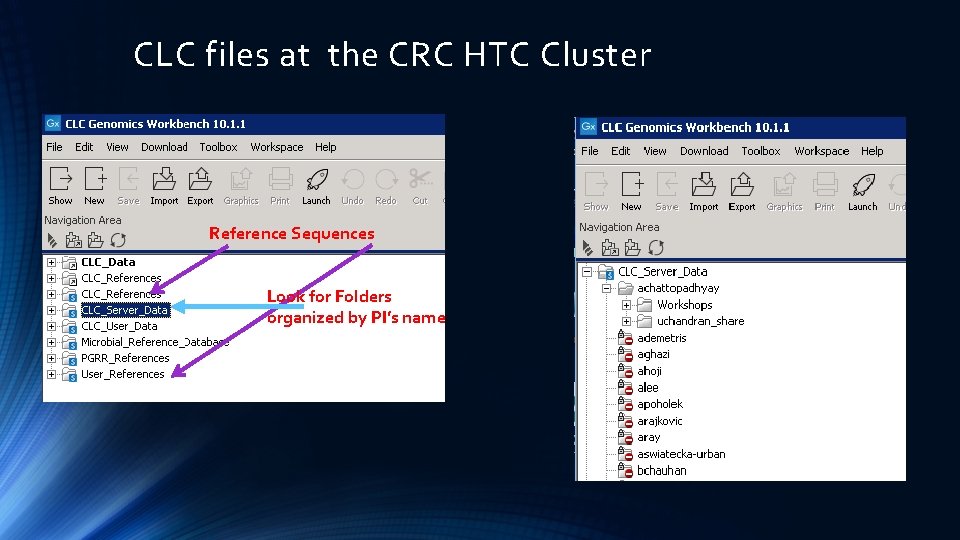
CLC files at the CRC HTC Cluster Reference Sequences Look for Folders organized by PI’s name
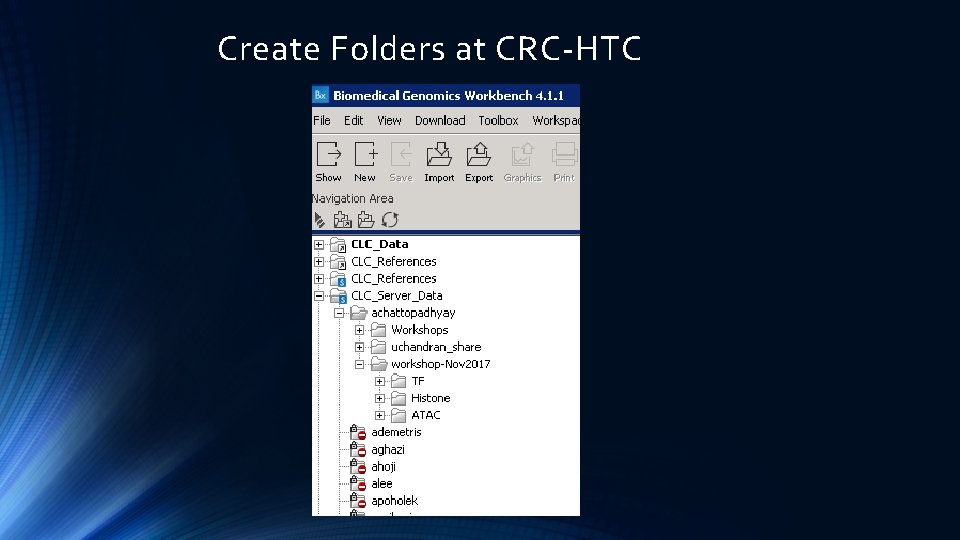
Create Folders at CRC-HTC
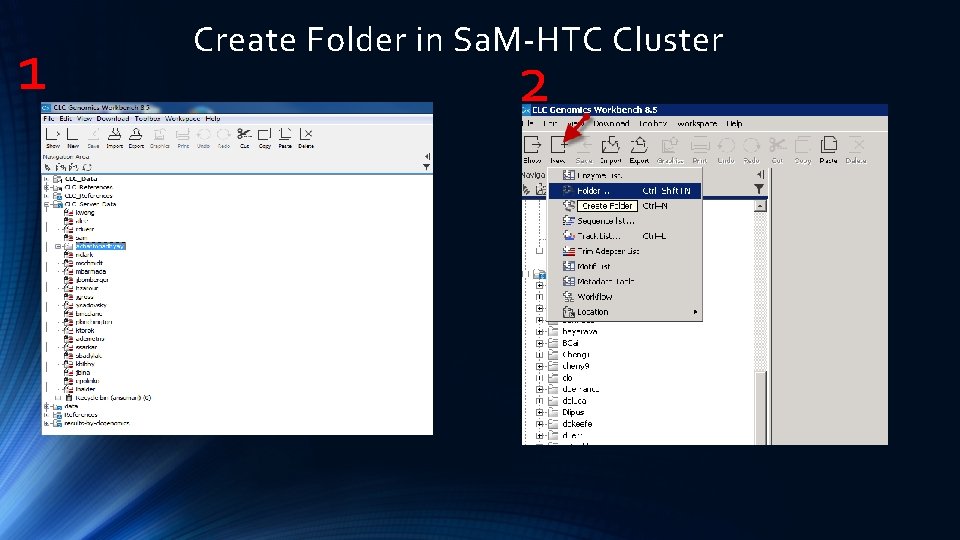
1 Create Folder in Sa. M-HTC Cluster 2
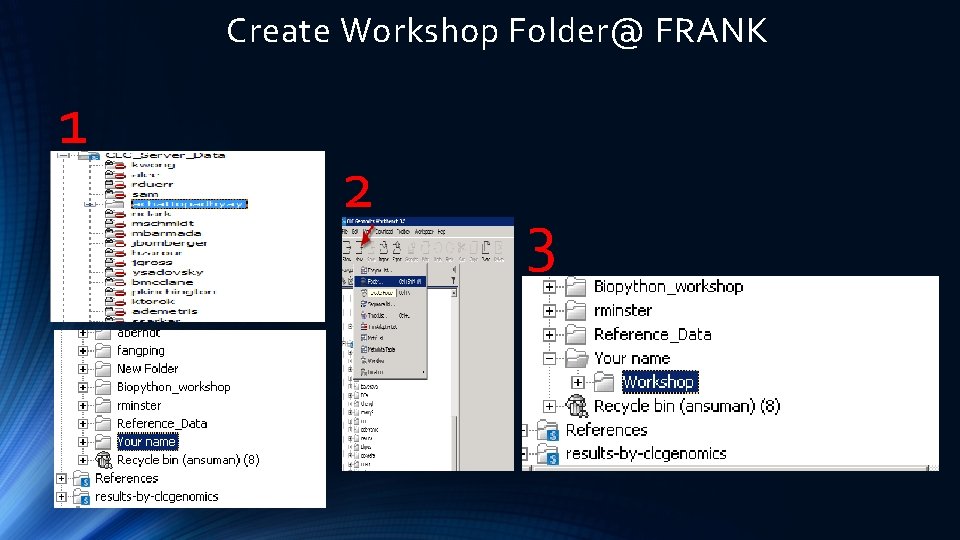
Create Workshop Folder@ FRANK 1 2 3
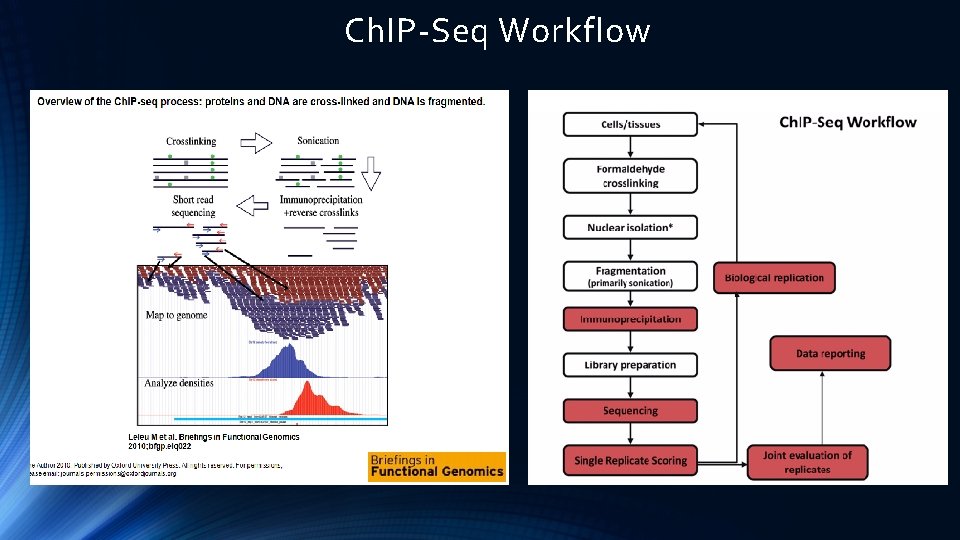
Ch. IP-Seq Workflow
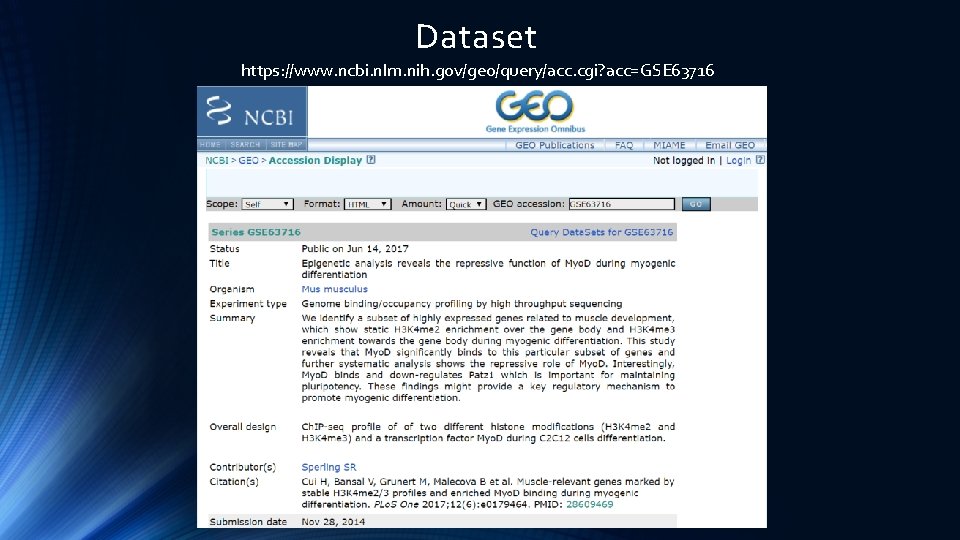
Dataset https: //www. ncbi. nlm. nih. gov/geo/query/acc. cgi? acc=GSE 63716
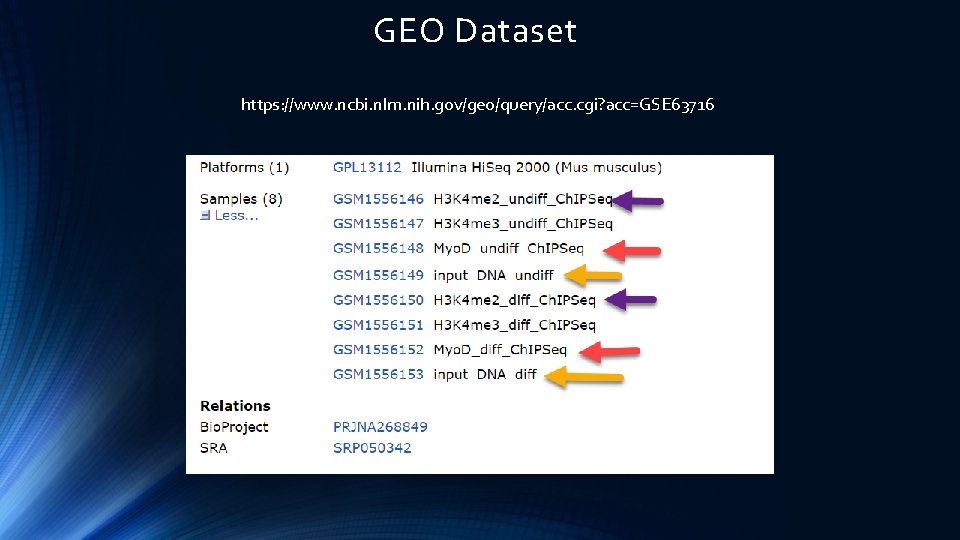
GEO Dataset https: //www. ncbi. nlm. nih. gov/geo/query/acc. cgi? acc=GSE 63716

Download FASTQ Reads Myo. D_Undiff_Ch. IP-Seq
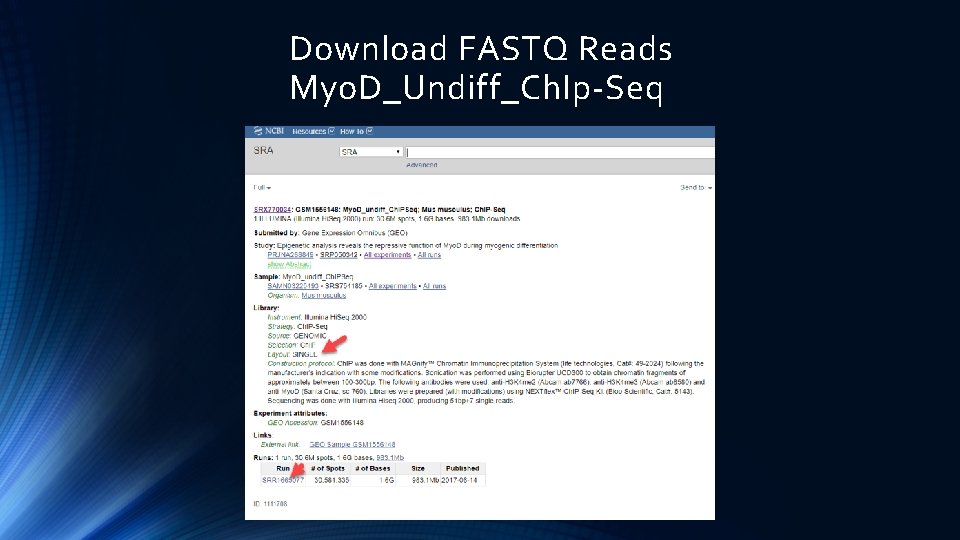
Download FASTQ Reads Myo. D_Undiff_Ch. Ip-Seq
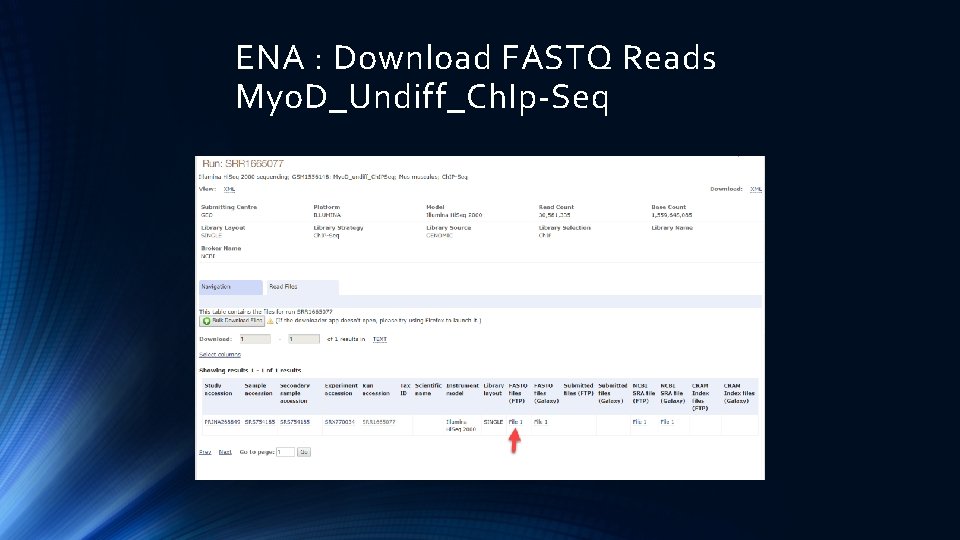
ENA : Download FASTQ Reads Myo. D_Undiff_Ch. Ip-Seq
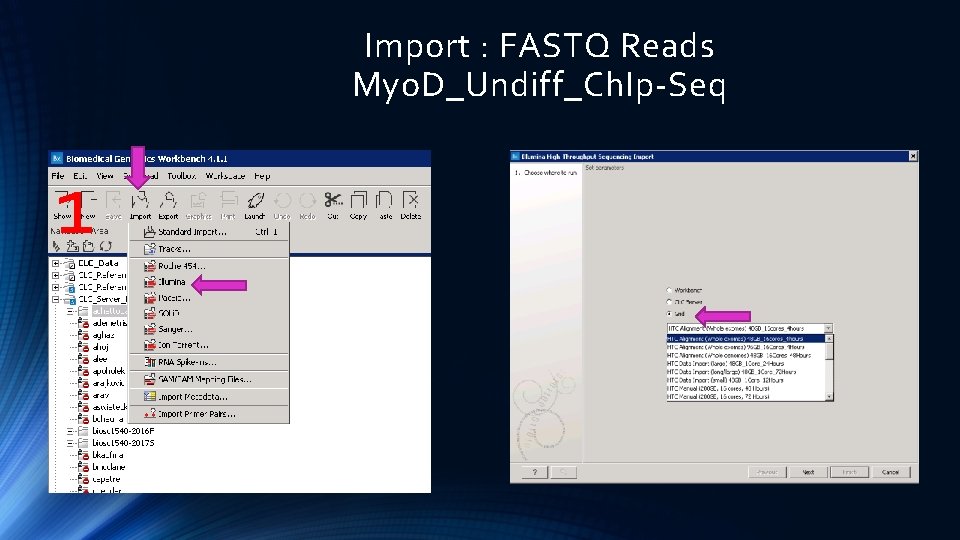
Import : FASTQ Reads Myo. D_Undiff_Ch. Ip-Seq 1
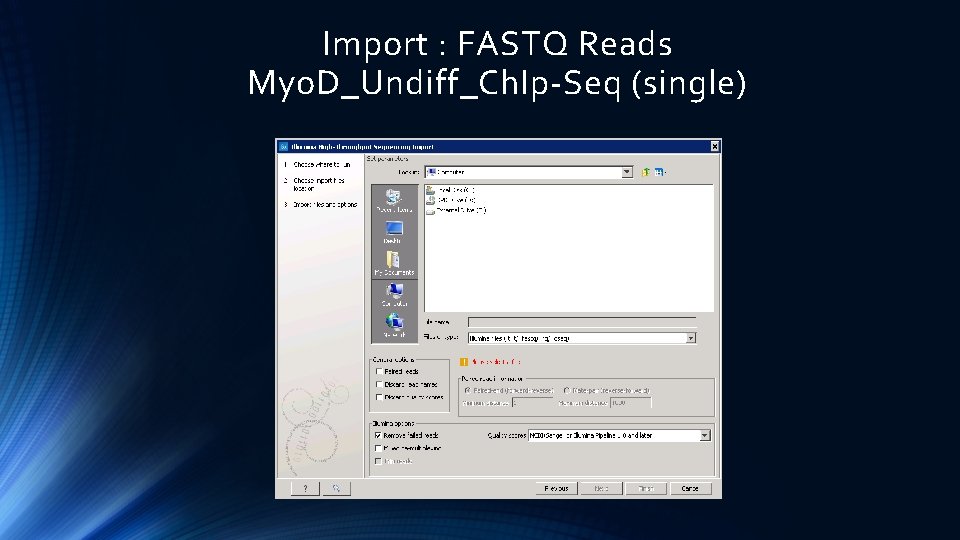
Import : FASTQ Reads Myo. D_Undiff_Ch. Ip-Seq (single)
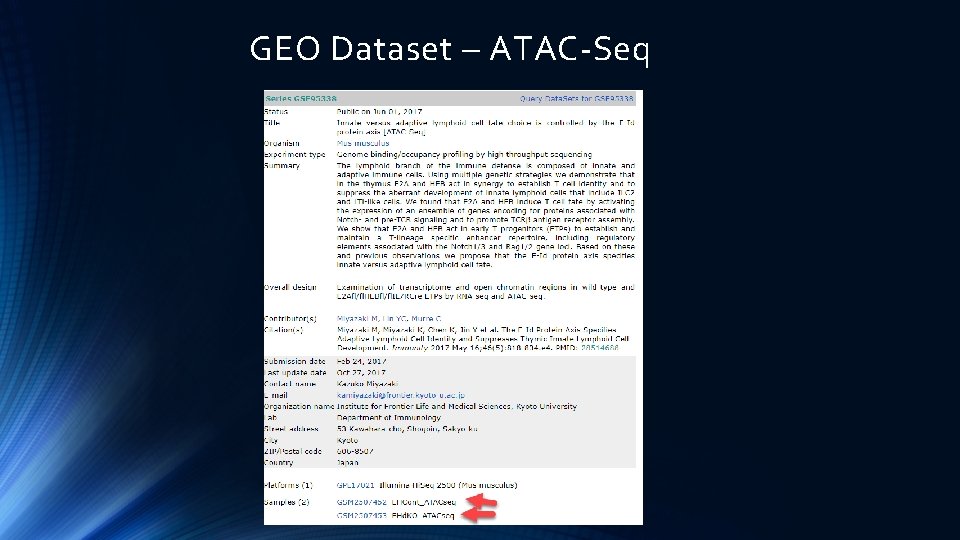
GEO Dataset – ATAC-Seq

1 STEP 1: Import Reads to CLC (Paired End) 2
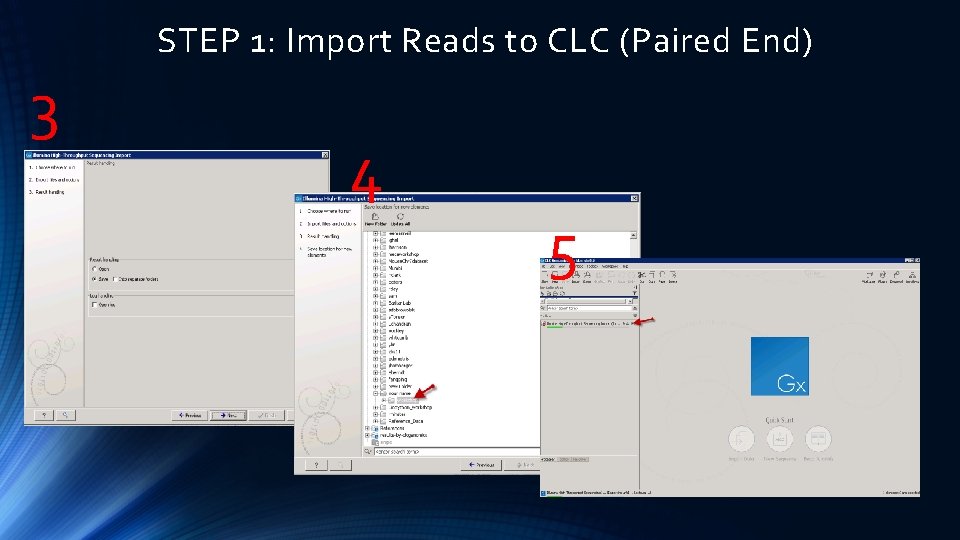
STEP 1: Import Reads to CLC (Paired End) 3 4 5
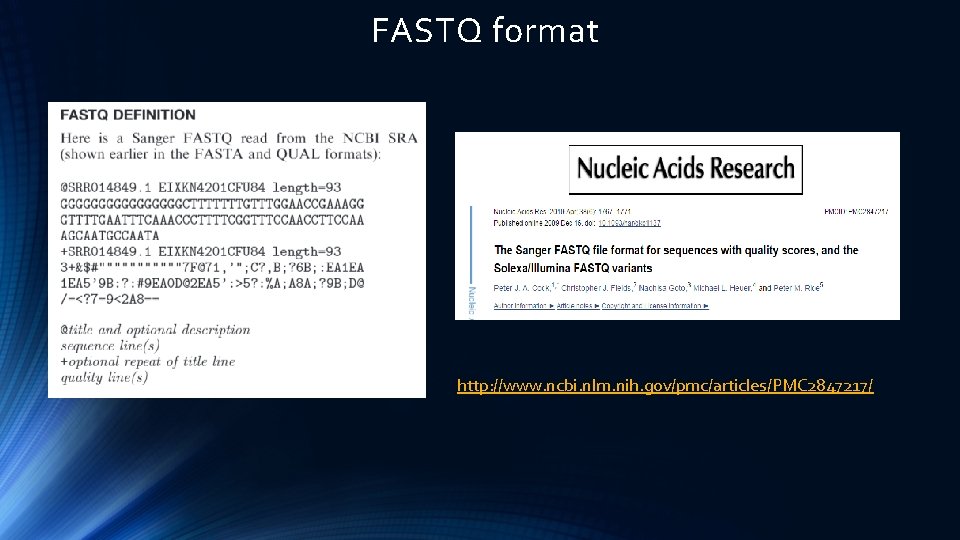
FASTQ format http: //www. ncbi. nlm. nih. gov/pmc/articles/PMC 2847217/

FASTQ Reads
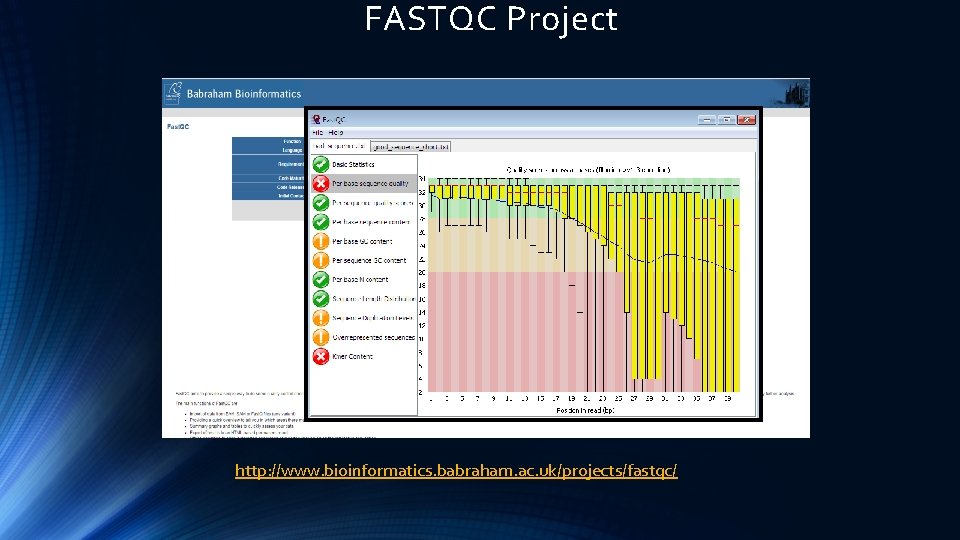
FASTQC Project http: //www. bioinformatics. babraham. ac. uk/projects/fastqc/
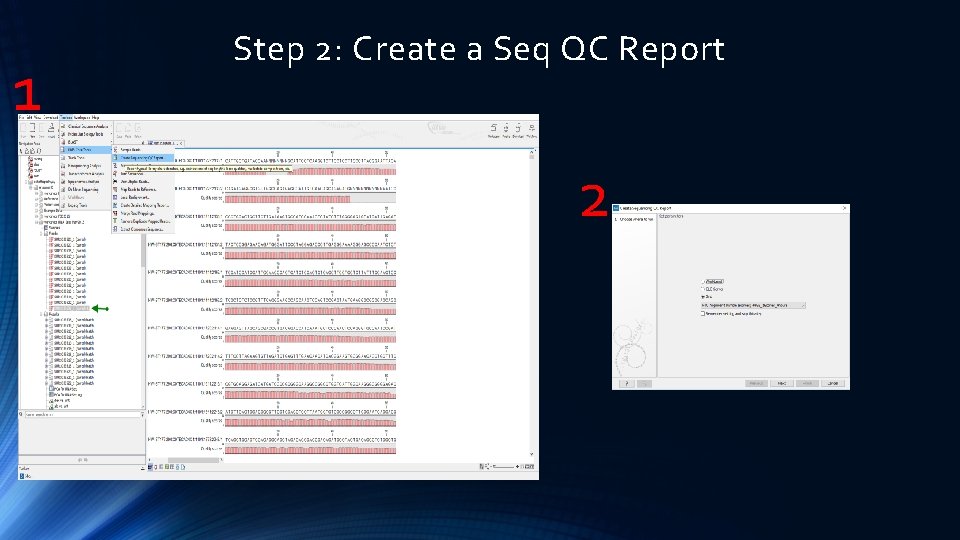
1 Step 2: Create a Seq QC Report 2
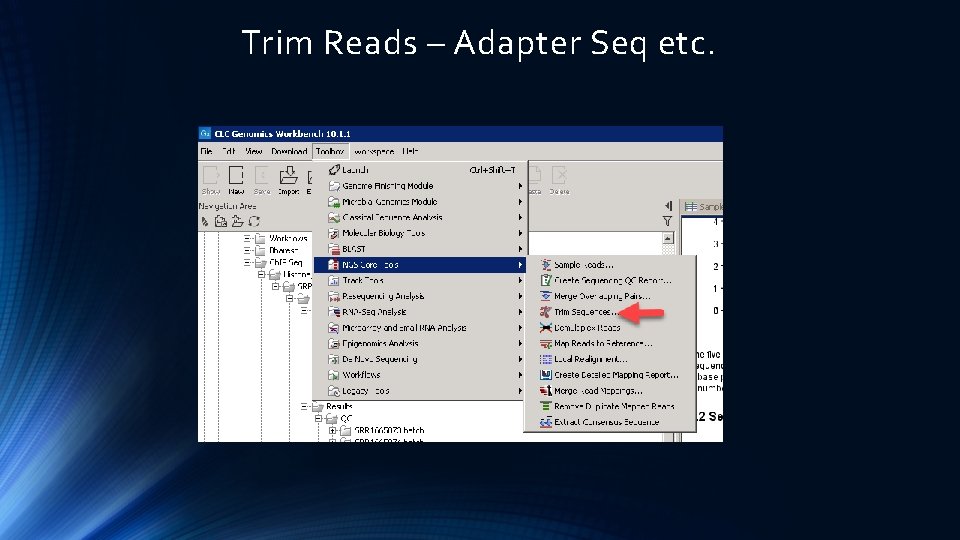
Trim Reads – Adapter Seq etc.
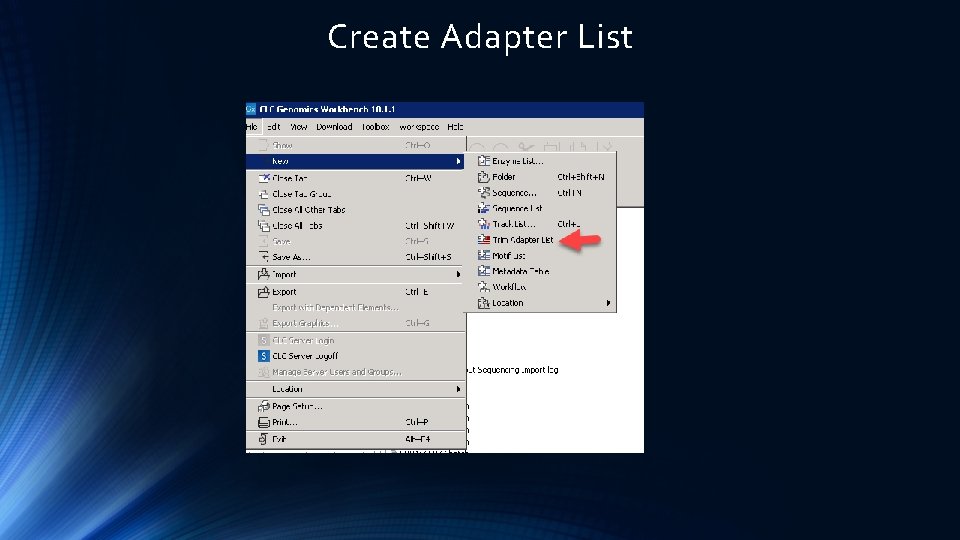
Create Adapter List
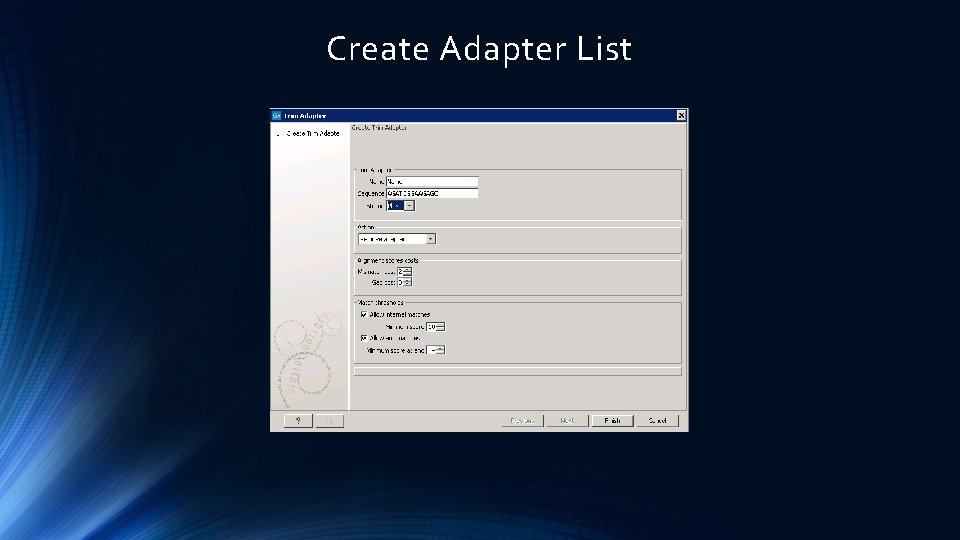
Create Adapter List
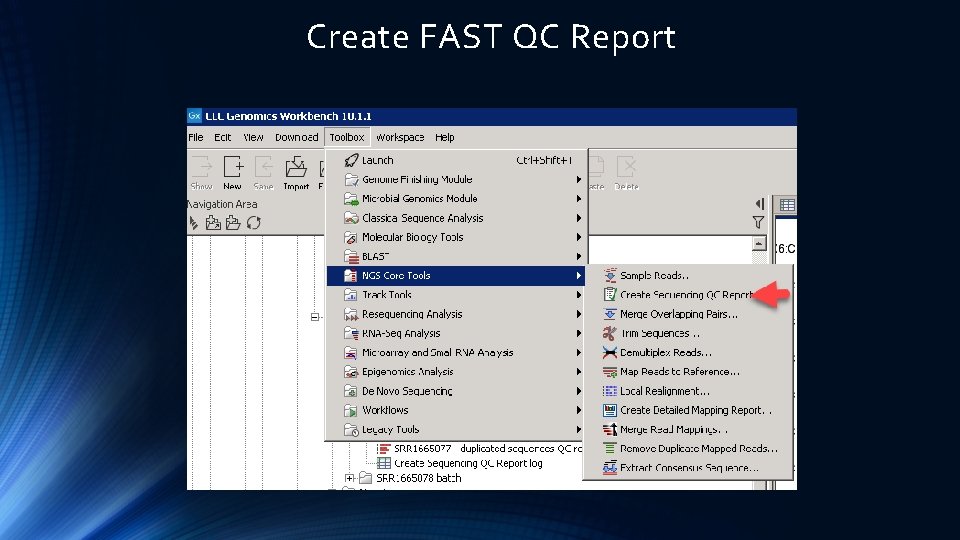
Create FAST QC Report
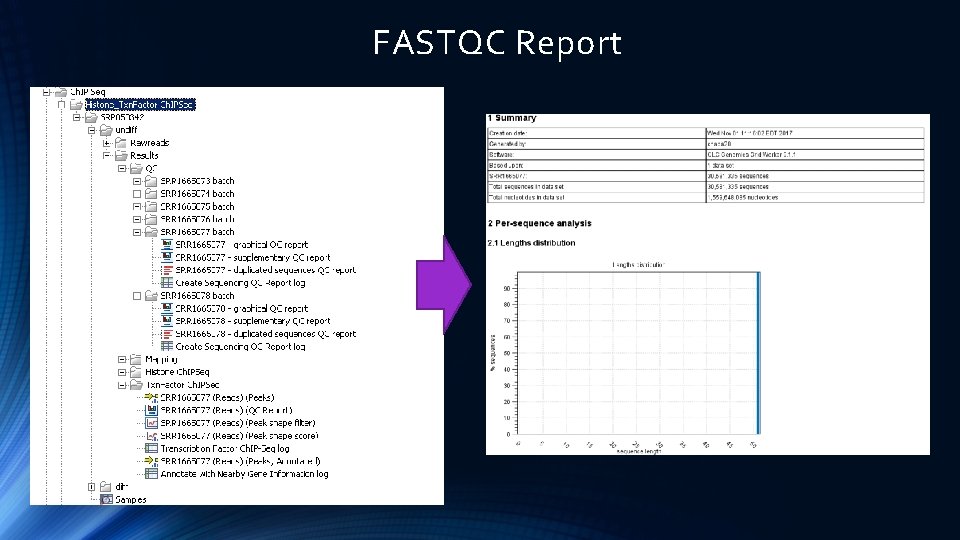
FASTQC Report
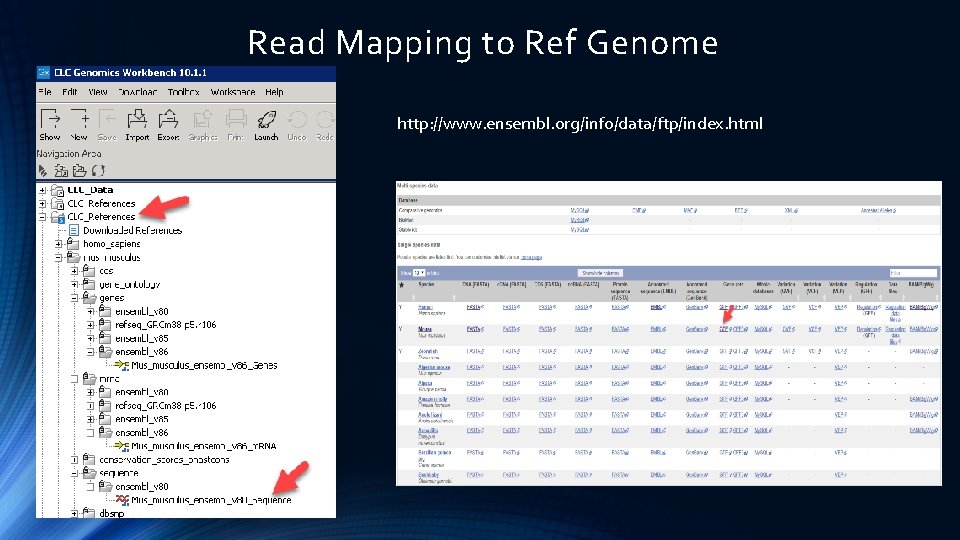
Read Mapping to Ref Genome http: //www. ensembl. org/info/data/ftp/index. html
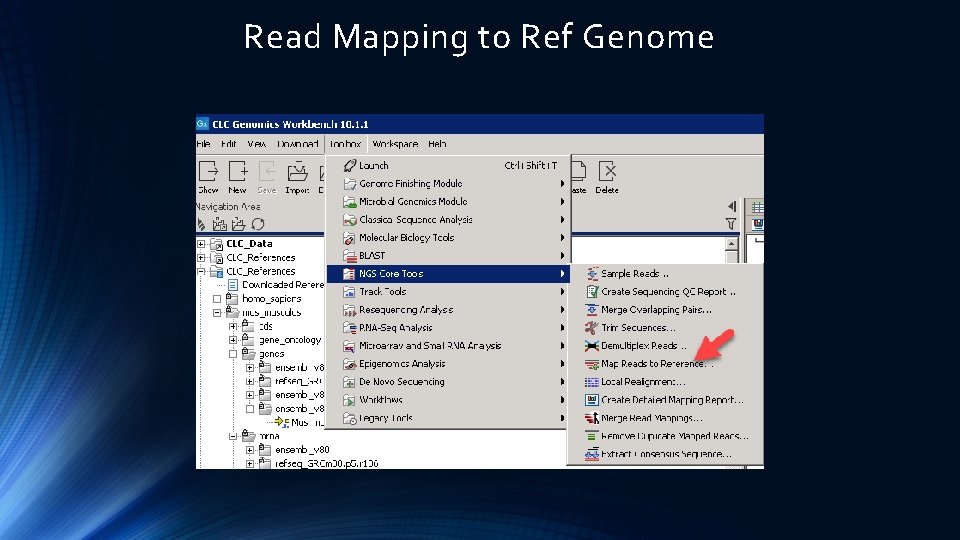
Read Mapping to Ref Genome
Read Mapping to Ref Genome
Read Mapping to Ref Genome

Read Mapping to Ref Genome
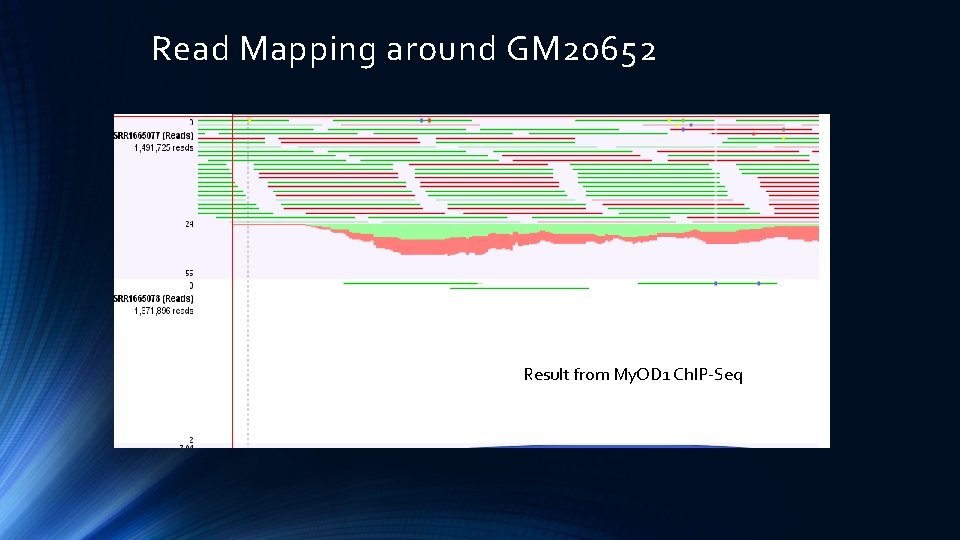
Read Mapping around GM 20652 Result from My. OD 1 Ch. IP-Seq
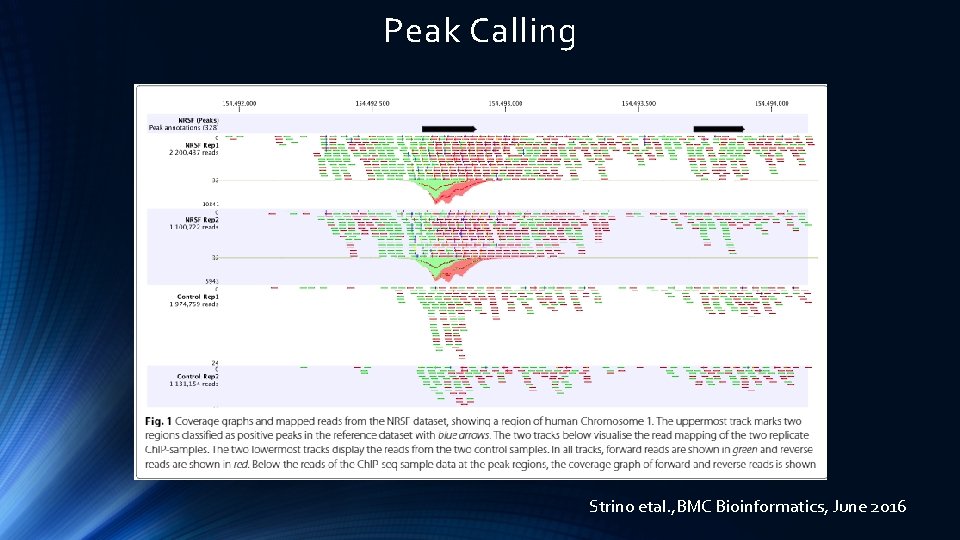
Peak Calling Strino etal. , BMC Bioinformatics, June 2016
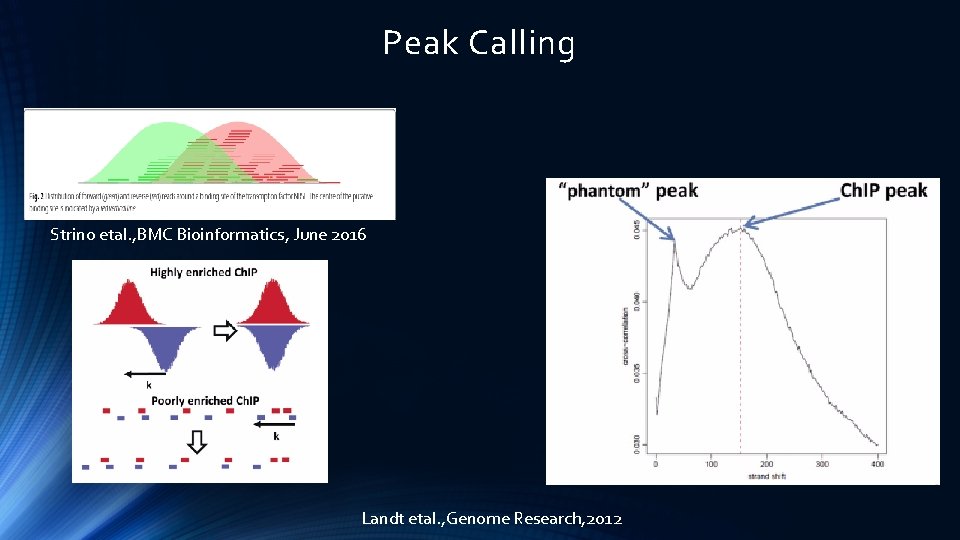
Peak Calling Strino etal. , BMC Bioinformatics, June 2016 Landt etal. , Genome Research, 2012

Peak Calling Strino etal. , BMC Bioinformatics, June 2016
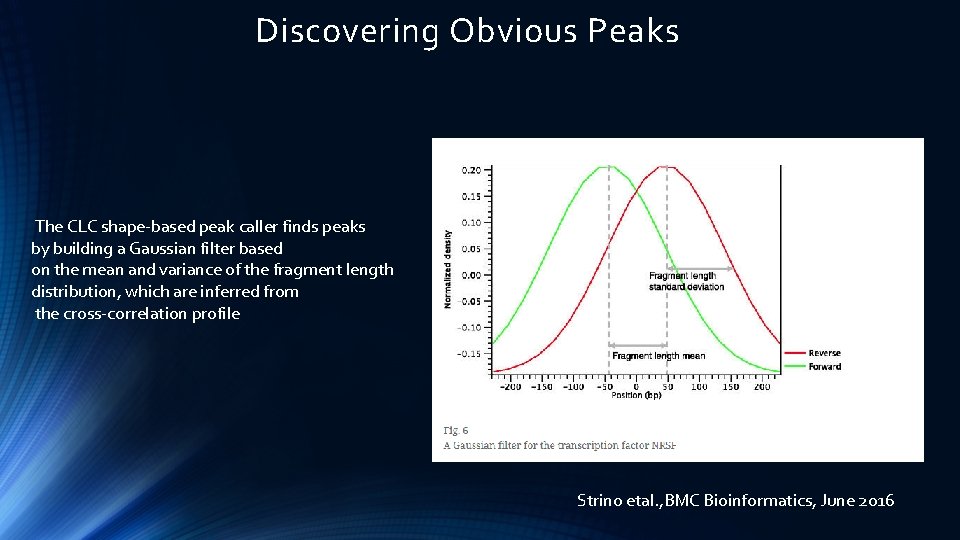
Discovering Obvious Peaks The CLC shape-based peak caller finds peaks by building a Gaussian filter based on the mean and variance of the fragment length distribution, which are inferred from the cross-correlation profile Strino etal. , BMC Bioinformatics, June 2016

Peak Shape Score The Peak Shape Score is standardised and follows a standard normal distribution, so a p-value for each genomic position can be calculated as p-value=Φ(−Peak Shape Score of the peak centre), where Φ is the standard normal cumulative distribution function. Score = genomic coverage * filter; *: cross-correlation operator Score indicates how likely a genomic position is to be a center of a peak Strino etal. , BMC Bioinformatics, June 2016

Peak Shape Filter Once the positive and negative regions have been identified, the CLC shape-based peak caller learns a filter that matches the average peak shape, which is called Peak Shape Filter. Strino etal. , BMC Bioinformatics, June 2016
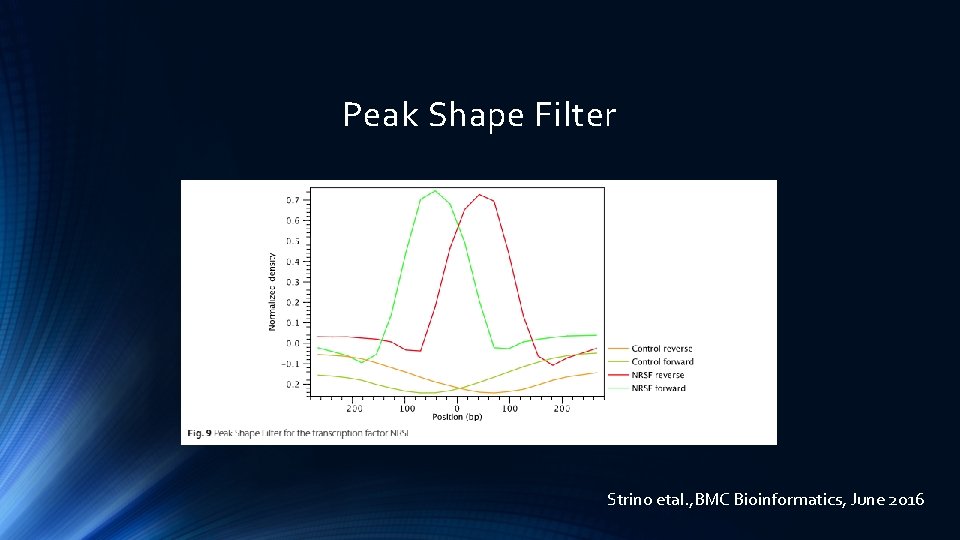
Peak Shape Filter Strino etal. , BMC Bioinformatics, June 2016
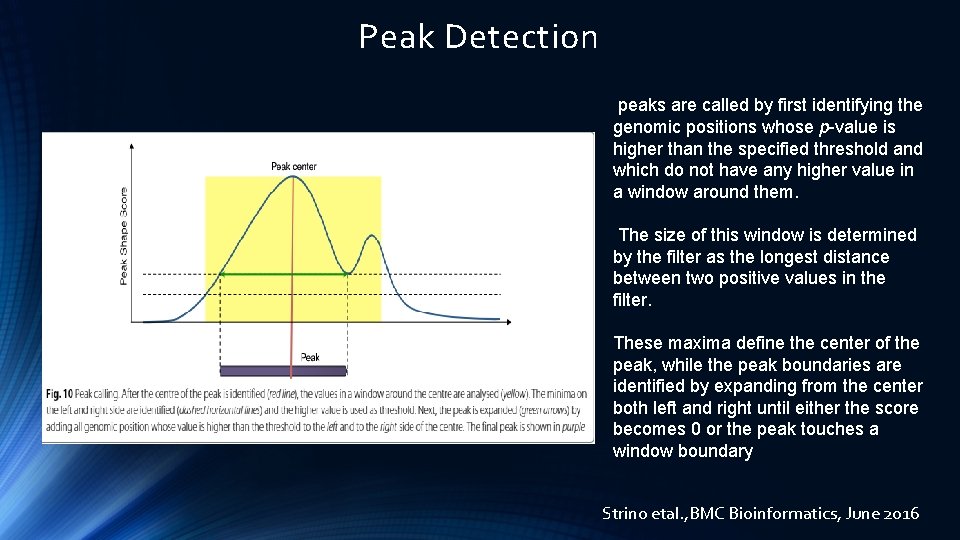
Peak Detection peaks are called by first identifying the genomic positions whose p-value is higher than the specified threshold and which do not have any higher value in a window around them. The size of this window is determined by the filter as the longest distance between two positive values in the filter. These maxima define the center of the peak, while the peak boundaries are identified by expanding from the center both left and right until either the score becomes 0 or the peak touches a window boundary Strino etal. , BMC Bioinformatics, June 2016
Call Peaks using Peak Shape information
Call Peaks using Peak Shape information
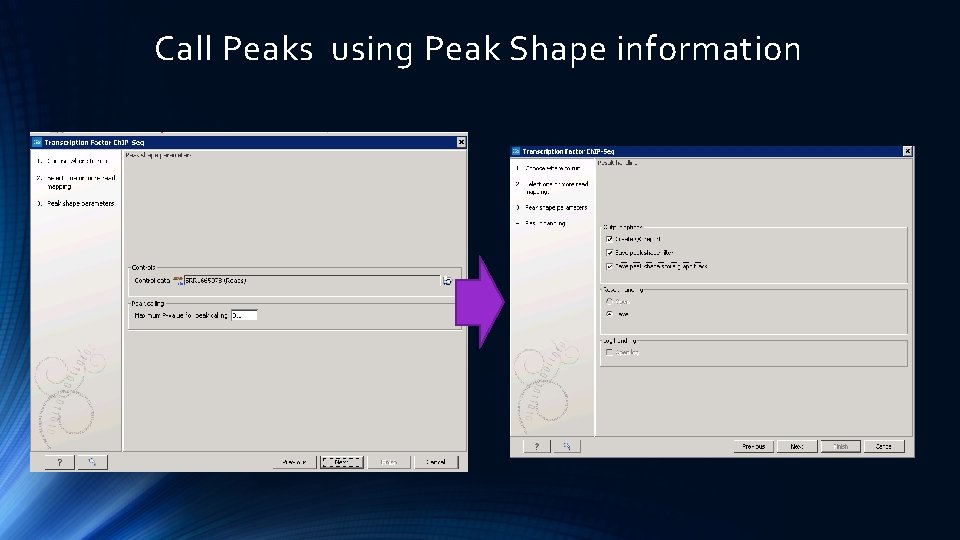
Call Peaks using Peak Shape information

Peak Calls Result
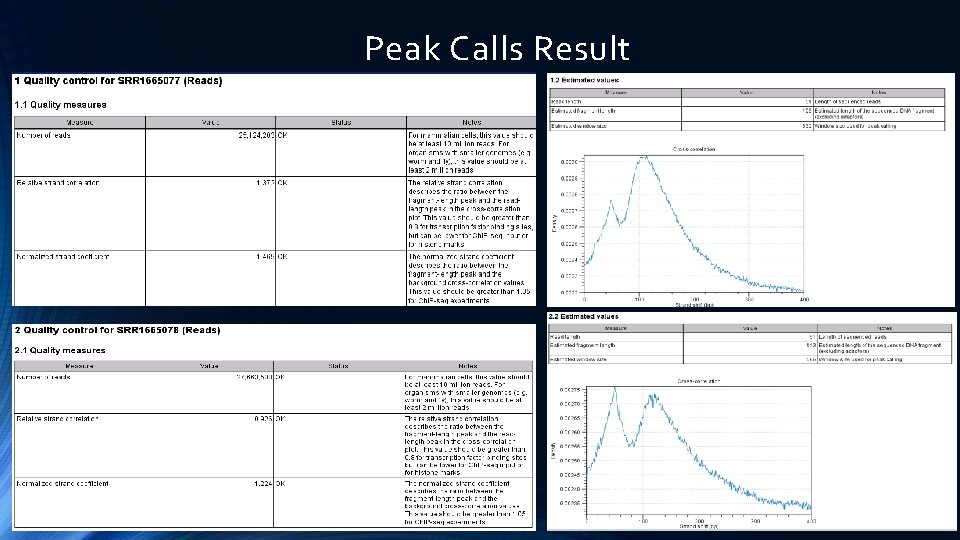
Peak Calls Result
Annotate Peaks with near by genes
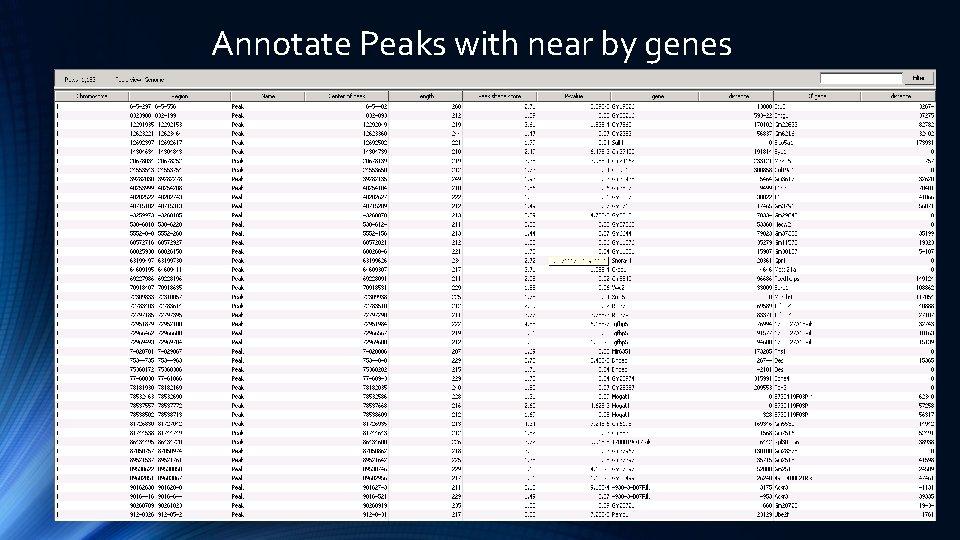
Annotate Peaks with near by genes

5 Prime and 3 Prime Gene Distance
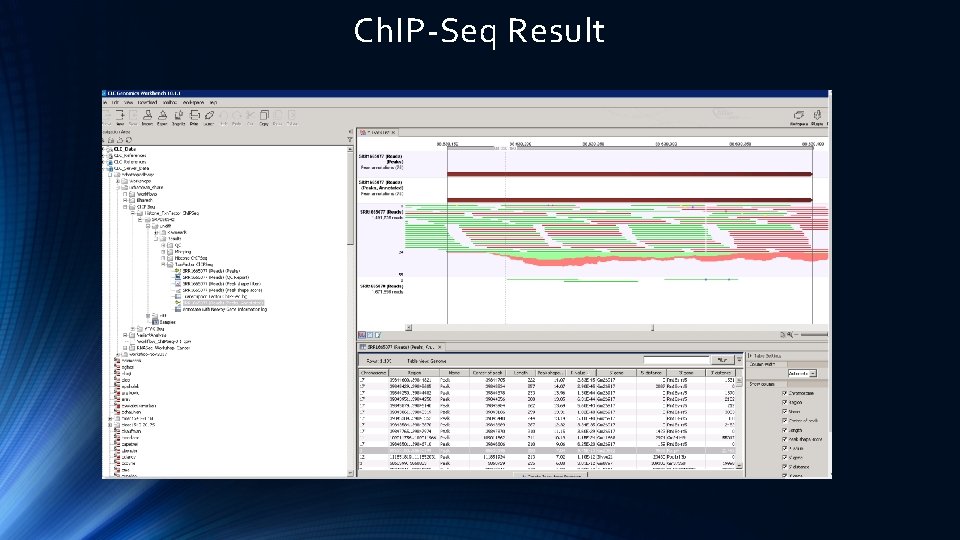
Ch. IP-Seq Result
Compare Datasets
Compare Datasets
Compare Datasets

Compare Datasets
Commonly Used Open-Source Tool https: //pypi. python. org/pypi/MACS 2
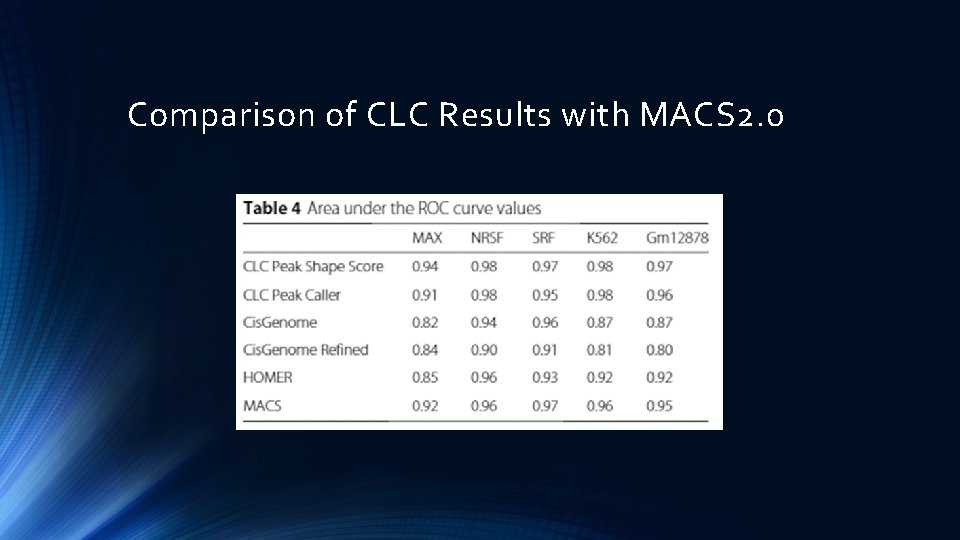
Comparison of CLC Results with MACS 2. 0
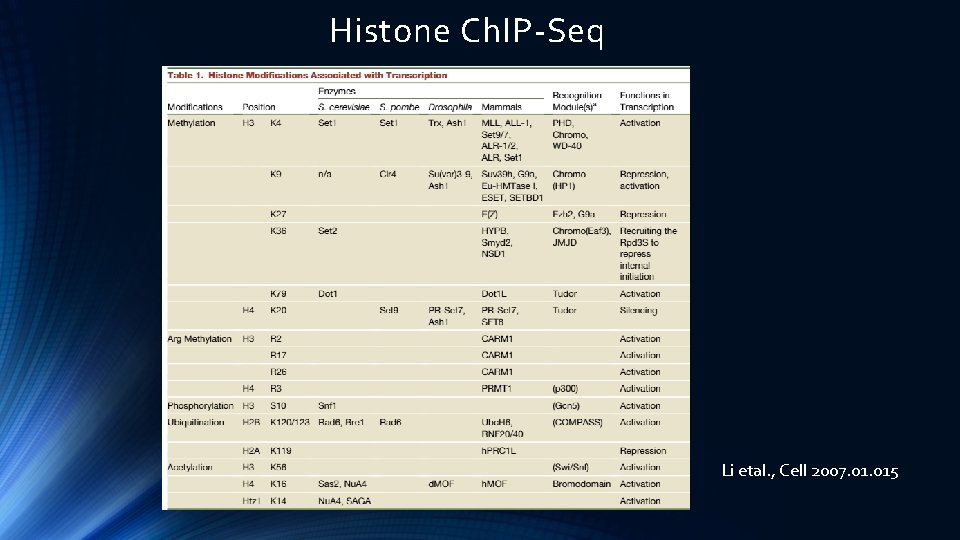
Histone Ch. IP-Seq Li etal. , Cell 2007. 015

Histone Ch. IP-Seq
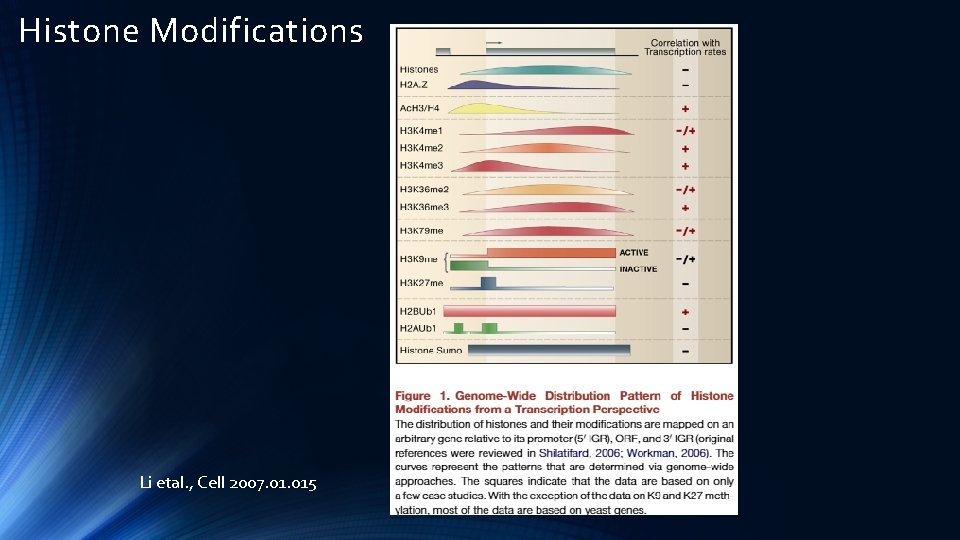
Histone Modifications Li etal. , Cell 2007. 015
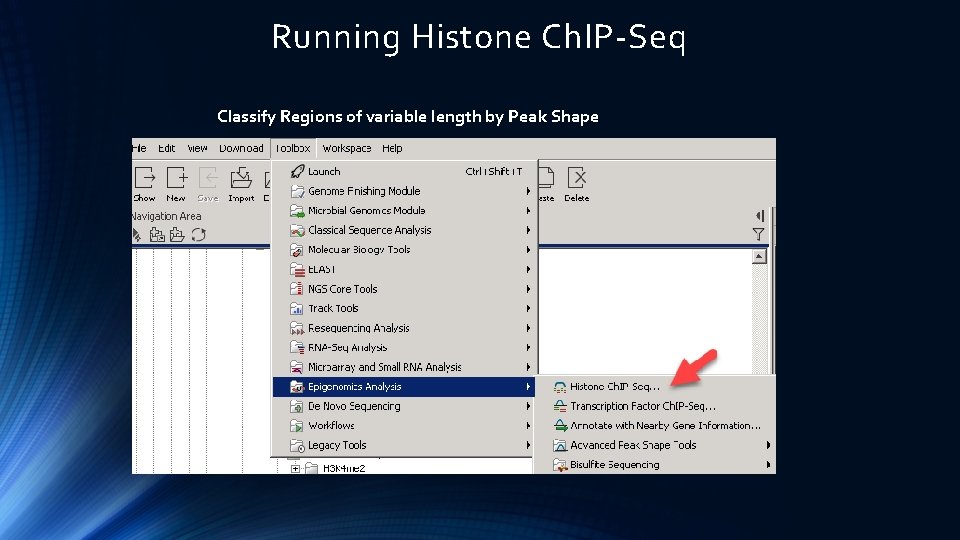
Running Histone Ch. IP-Seq Classify Regions of variable length by Peak Shape

Running Histone Ch. IP-Seq
Running Histone Ch. IP-Seq
Running Histone Ch. IP-Seq
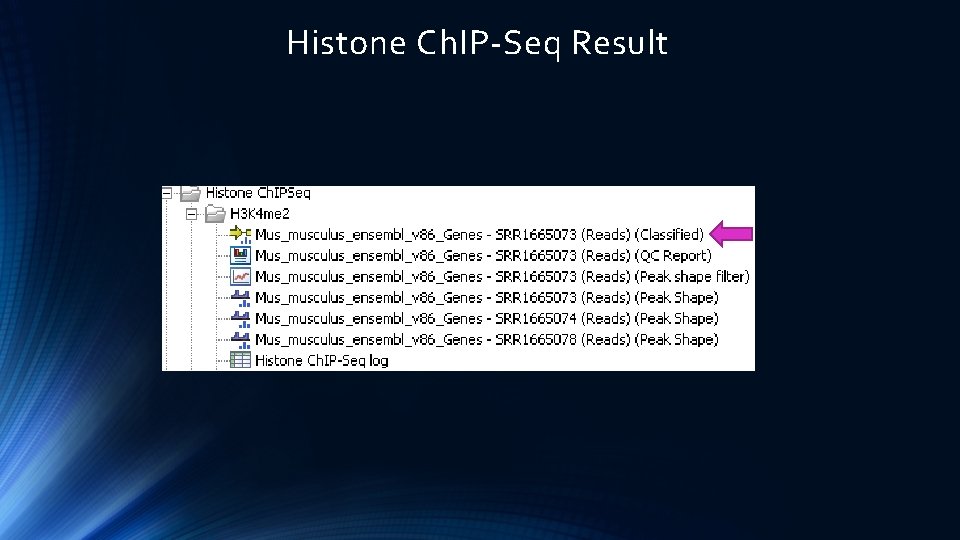
Histone Ch. IP-Seq Result
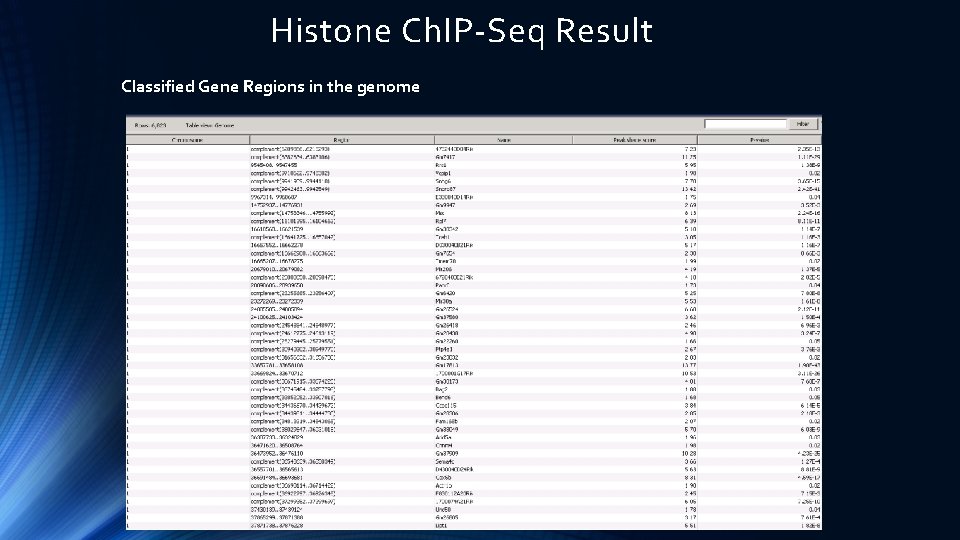
Histone Ch. IP-Seq Result Classified Gene Regions in the genome

H 3 K 4 Me 3 – Diff : Result by Txnfactor Ch. IPSeq tool
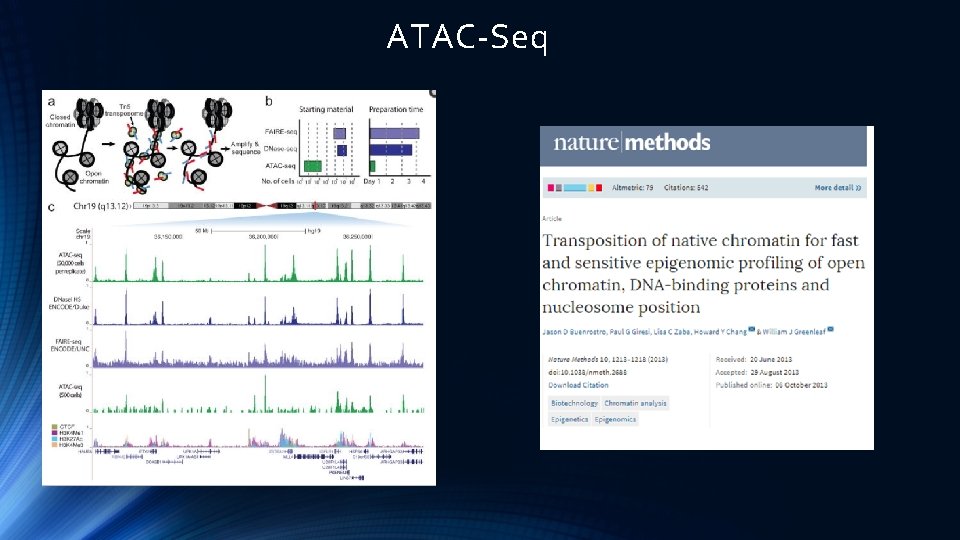
ATAC-Seq

ATAC-Seq Data Analysis
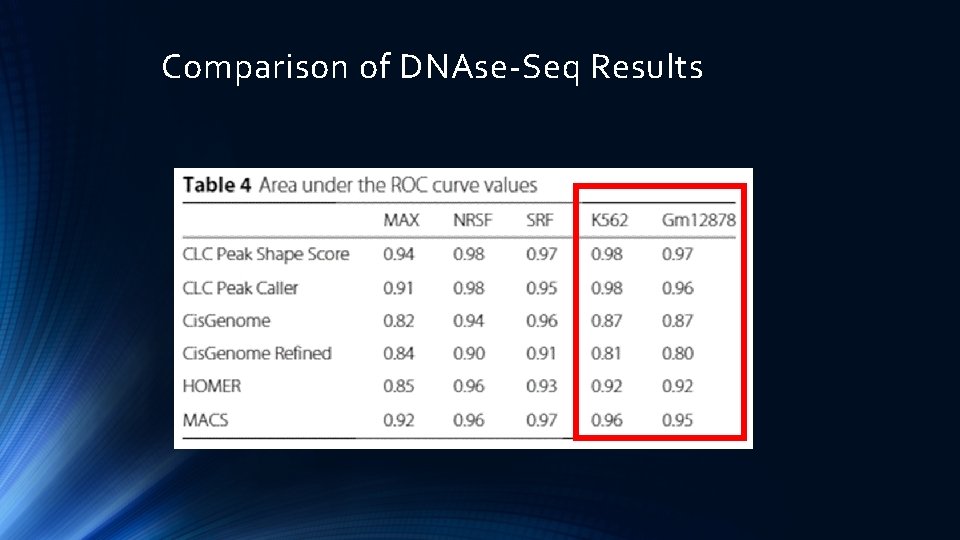
Comparison of DNAse-Seq Results

HSLS-MBIS and Genomics Analysis Core HSLS-MBIS GAC Uma Chandran, Ph. D, MSIS 412 -648 -9326 Chandran@pitt. edu Ansuman Chattopadhyay, Ph. D 412 -648 -1297 ansuman@pitt. edu Sri Chaparala srichaparala@pitt. edu Carrie Iwema, Ph. D, MLS 412 -383 -6887 iwema@pitt. edu http: //hscrf. pitt. edu/

Thanks To…. • HSLS Sri Chaparala Carrie Iwema David Leung Michael Sweezer • CLCBio Shawn Prince • Center for Research Computing Mu Fangping
- Slides: 80