Analyzing Ch IPseq data Running MACS and MEME





![MACS usage Usage: macs 2 <-t tfile> [-n name] [ -g genomesize] [options] Example: MACS usage Usage: macs 2 <-t tfile> [-n name] [ -g genomesize] [options] Example:](https://slidetodoc.com/presentation_image_h/af3b29ffee08688ab6005a4a48107422/image-6.jpg)








- Slides: 18

Analyzing Ch. IP-seq data Running MACS and MEME

Prepare your directory for today’s class 1. Remove any un-needed files from your home directory 2. Make directory & subdirectories for today

Add files for today’s class 1. cp /projects/sreadgrp/Ch. IP_class/CTCF_ chr 21. sorted. bam* ~/Ch. IP/SAM/ 2. cp /projects/sreadgrp/Ch. IP_class/CTCF_ input_chr 21. sorted. bam* ~/Ch. IP/SAM/ 3. cp /projects/sreadg rp/Ch. IP_class/PBS_scripts/* ~/Ch. IP/PBS/

Load modules 1. module load MACS_2. 0. 9 2. module load meme_4. 8. 1

Using MACS
![MACS usage Usage macs 2 t tfile n name g genomesize options Example MACS usage Usage: macs 2 <-t tfile> [-n name] [ -g genomesize] [options] Example:](https://slidetodoc.com/presentation_image_h/af3b29ffee08688ab6005a4a48107422/image-6.jpg)
MACS usage Usage: macs 2 <-t tfile> [-n name] [ -g genomesize] [options] Example: macs 2 -t Ch. IP. bam -c Control. bam -f BAM -g hs -n test -B -q 0. 01 or example for broad peak calling: macs 2 -t Ch. IP. bam -c Control. bam -broad -g hs

Edit MACS_CTCF. pbs 1. Replace <___> with appropriate information 2. Replace “amso 5880” with identikey

Copy files to /projects/sreadgrp/student/identikey/ $ cp /Users/identikey/Ch. IP/BAM/CTCF_chr 2 1. sorted. bam* /projects/sreadgroup/student/identi key/ $ cp /Users/Identikey/Ch. IP/MACS_out/CTCF _chr 21_peaks. bed /projects/sreadgroup/student/identi

Visualize Ch. IP data 1. Start visualization & open IGV 2. Load IGV genome file from: /projects/sreadgrp/student/IGV_geno mes/Hg 19_Chr 21_igv. genome 3. Open sorted. bam files 4. Open peaks. bed file

Using MEME

Getting to know MEME $ meme –help 1. 2. 3. 4. What type of file does MEME take in? What does MEME output? What does the –dna option mean? How do you set motif width?

Make fasta from bed • Edit Bedto. Fasta_CTCF. pbs 1. Replace <___> appropriately 2. Replace “amso 5880” with identikey

Edit MEME_CTCF. pbs 1. Replace <___> appropriately 2. Replace “amso 5880” with identikey

Copy files to visualization folder $ cp /Users/identikey/Ch. IP/ctcf_meme_out /* /projects/sreadgroup/student/identi key/

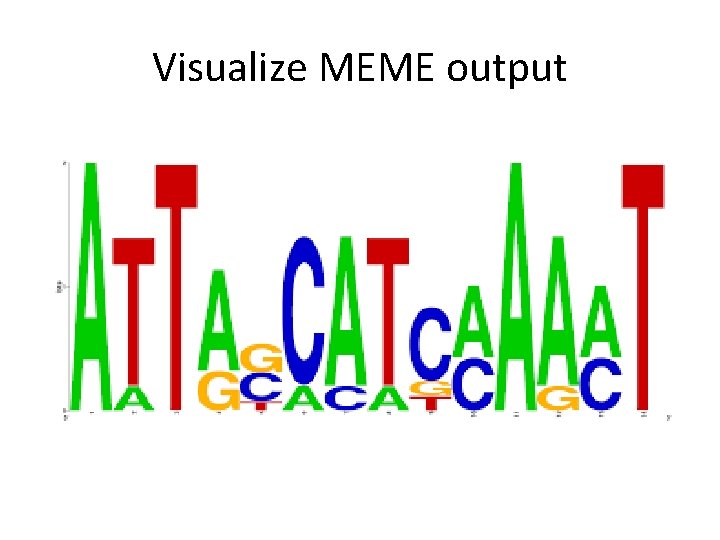
Visualize MEME output

MEME Suite $ ls /opt/meme/4. 8. 1/bin/

MEME suite online http: //meme. ebi. edu. au/meme/index. html

Suggestions for MEME • Try using DREME for short motifs, or if you are having difficulty getting good results from MEME • Rerun MEME with shorter motif • Downstream analysis: – FIMO – MAST