Cell Designer Tutorial Laurence Calzone Andrei Zinovyev UMR




![Start with a simple network - Open a new document: [File] => [New] - Start with a simple network - Open a new document: [File] => [New] -](https://slidetodoc.com/presentation_image_h/dfd16326367e4209811037030ff38f52/image-20.jpg)

![Some functions Drawing tips: - Choose from the [Edit] menu, Grid Snap, to help Some functions Drawing tips: - Choose from the [Edit] menu, Grid Snap, to help](https://slidetodoc.com/presentation_image_h/dfd16326367e4209811037030ff38f52/image-27.jpg)


- Slides: 34

Cell. Designer Tutorial Laurence Calzone, Andrei Zinovyev UMR U 900 INSERM/Institut Curie/Ecole des Mines de Paris Wednesday, April 30 th

Outline • Cell. Designer • Tutorial - Practice • Example

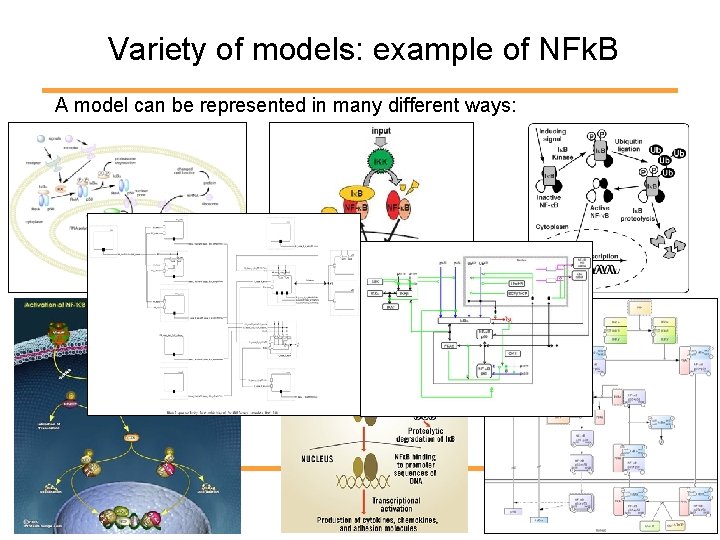
Variety of models: example of NFk. B A model can be represented in many different ways:

Many representations for the same model. => Each modeler uses its own representation => Need to read the paper to understand the model => Before being able to use or translate models into mathematics, need to understand the modeler’s symbols => Need for a standard format to be able to - use models - exchange models - compare models - compose models - etc.

Standard formats SBML (Systems Biology Markup Language) format for representing models of biochemical reaction networks Over 100 software packages support SBML Bio. PAX (Biological PAthway e. Xchange) format for storing biological pathway data; ontology for describing biochemical processes and their hierarchies SBGN (Systems Biology Graphical Notation) format for representing graphically biological interactions e. g. protein-protein interaction, gene regulatory networks, etc.

Cell. Designer: a short overview

What is Cell. Designer? Cell. Designer is a structured diagram editor for drawing gene-regulatory and biochemical networks that uses standard formats.

Cell. Designer content • • Full SBML support Graphical notation (SBGN) Possibility for mathematical simulations Database connection Export to PDF, PNG, … Freely available Supported Environment – – – Windows (2000 or later) Mac OS X Linux

What is Cell. Designer good for? Cell. Designer: • Offers an « easy-to-use » graphical representation of a network • Facilitates exchange of models developed by different groups, using a standard language (SBML: Systems Biology Markup Language) • Allows to annotate the reactions, proteins, genes, etc. • Proposes links to other databases (Pub. Med, IHOP, Bio. Models, KEGG, etc. ) • Organizes a lot of information in a unique diagram • Allows an exchange with other modeling tools

Install and Start Cell. Designer Download Cell. Designer version 4. 0 β from http: //celldesigner. org Download: Setup: Launch:

Start Cell. Designer

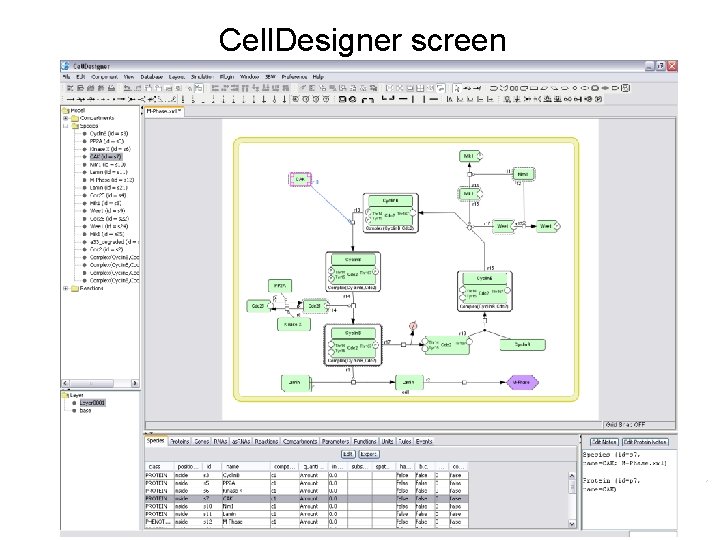
Cell. Designer screen

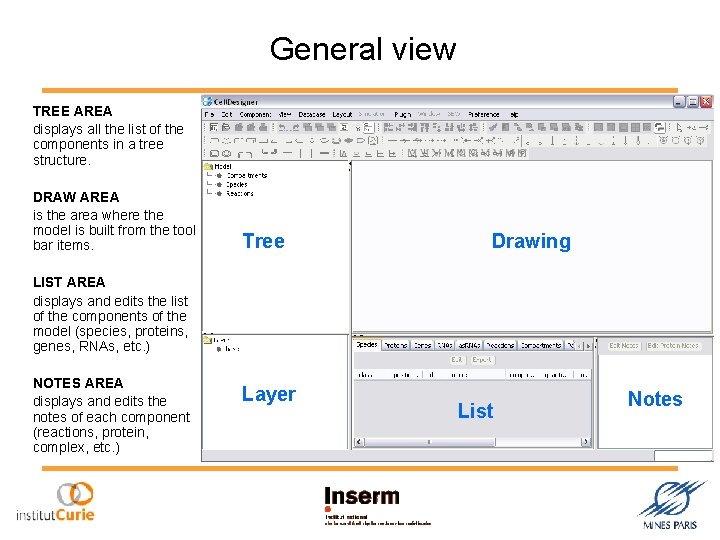
General view TREE AREA displays all the list of the components in a tree structure. DRAW AREA is the area where the model is built from the tool bar items. Tree Drawing LIST AREA displays and edits the list of the components of the model (species, proteins, genes, RNAs, etc. ) NOTES AREA displays and edits the notes of each component (reactions, protein, complex, etc. ) Layer List Notes

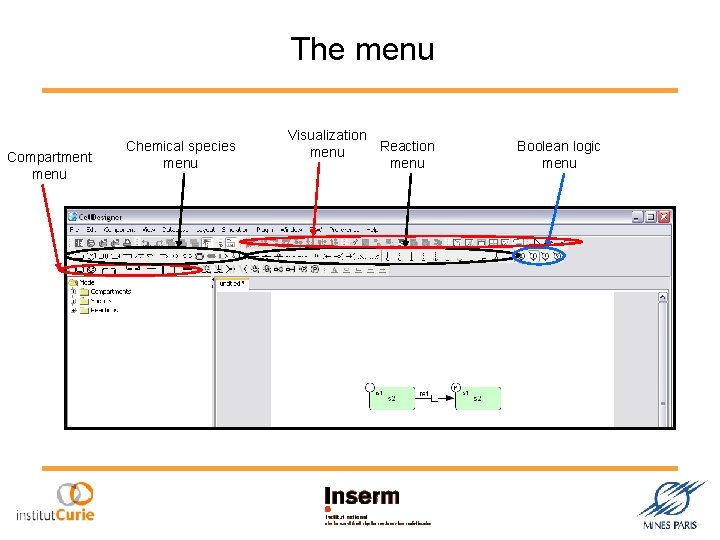
The menu Compartment menu Chemical species menu Visualization Reaction menu Boolean logic menu


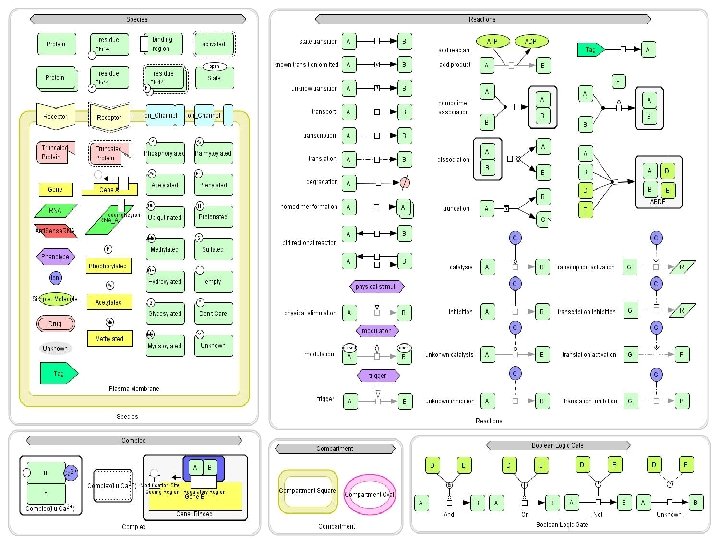
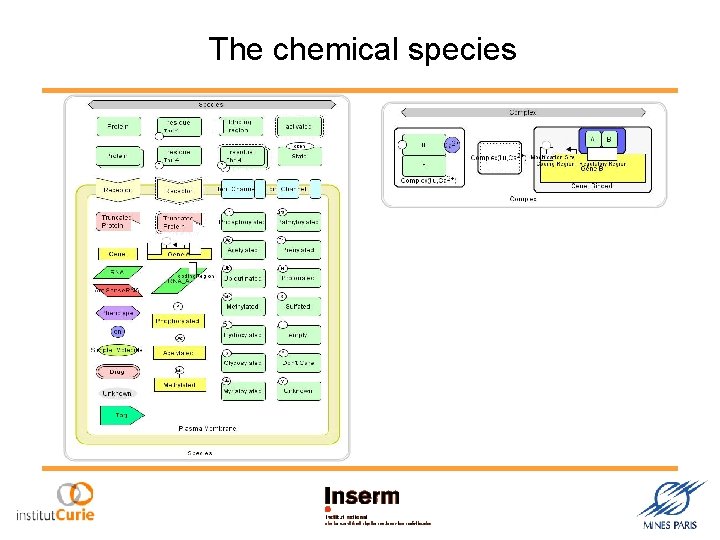
The chemical species

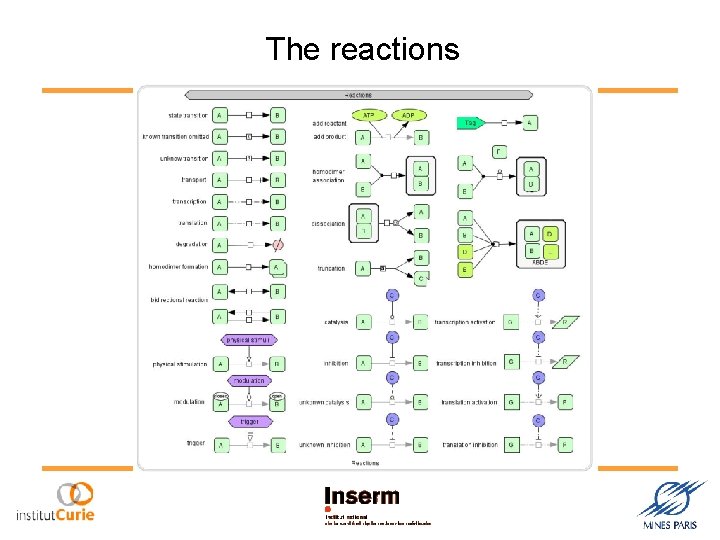
The reactions

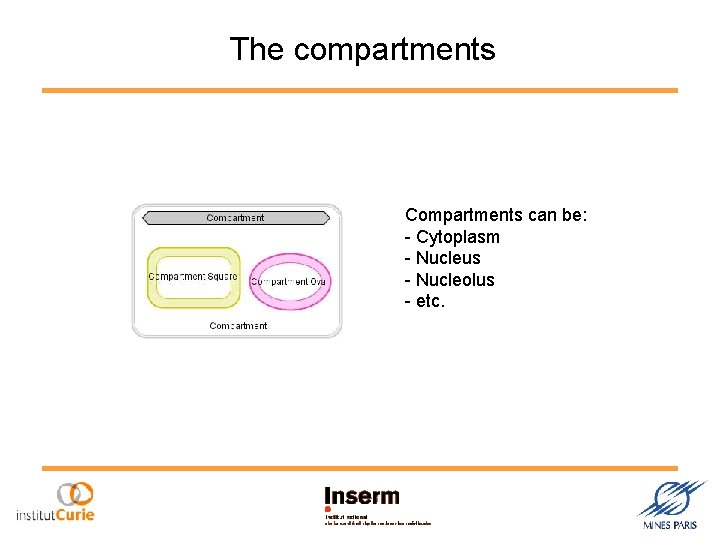
The compartments Compartments can be: - Cytoplasm - Nucleus - Nucleolus - etc.

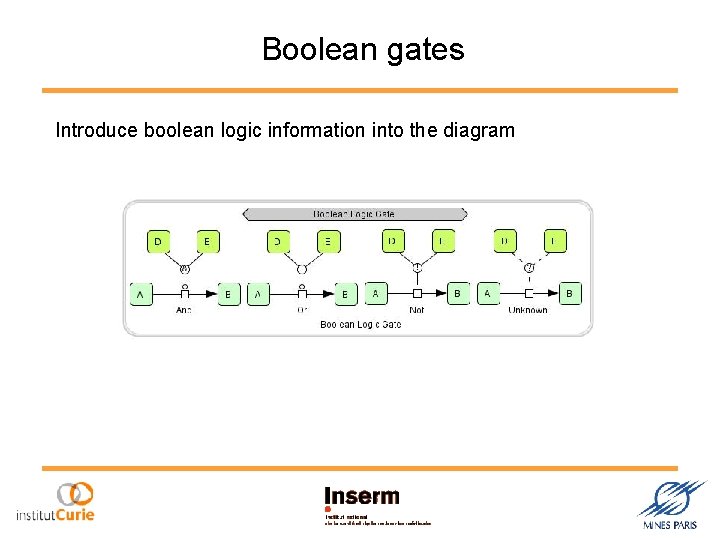
Boolean gates Introduce boolean logic information into the diagram
![Start with a simple network Open a new document File New Start with a simple network - Open a new document: [File] => [New] -](https://slidetodoc.com/presentation_image_h/dfd16326367e4209811037030ff38f52/image-20.jpg)
Start with a simple network - Open a new document: [File] => [New] - Name your network (Michaelis. Menten) - Choose the dimension of the graph (by default: 600 by 400 but can be changed later)

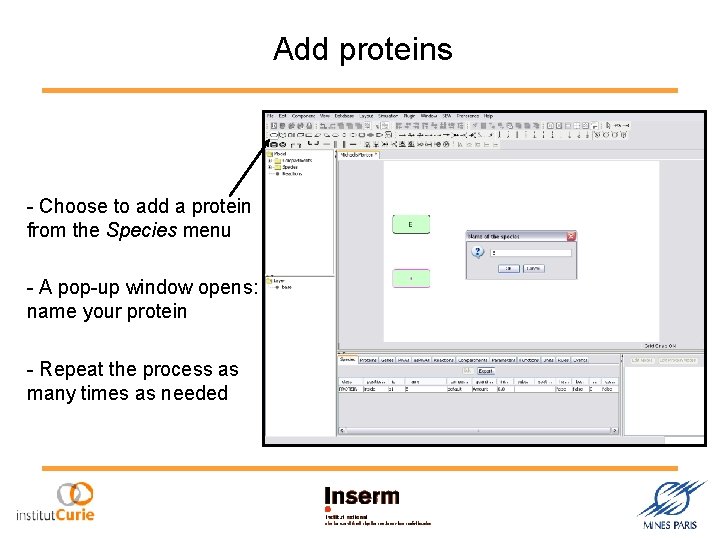
Add proteins - Choose to add a protein from the Species menu - A pop-up window opens: name your protein - Repeat the process as many times as needed

Add complexes - Choose to add a complex from the Species menu - Name it - Insert the proteins that compose the complexes (you must copy/paste the proteins from the diagram itself and move them inside the complex)

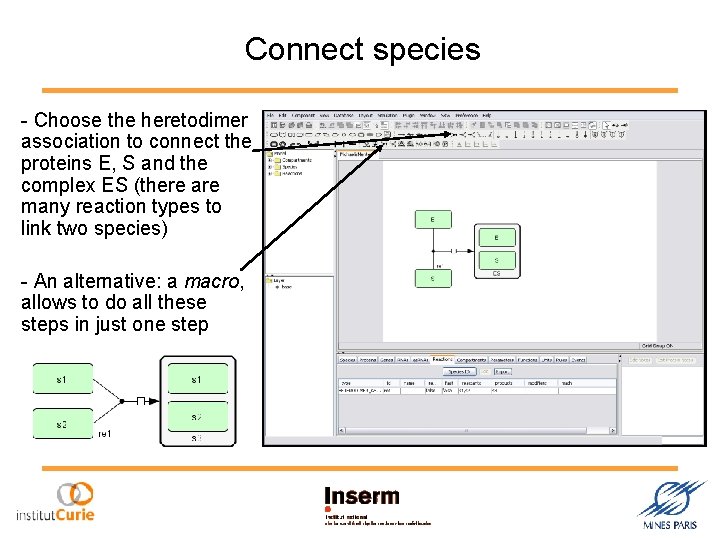
Connect species - Choose the heretodimer association to connect the proteins E, S and the complex ES (there are many reaction types to link two species) - An alternative: a macro, allows to do all these steps in just one step

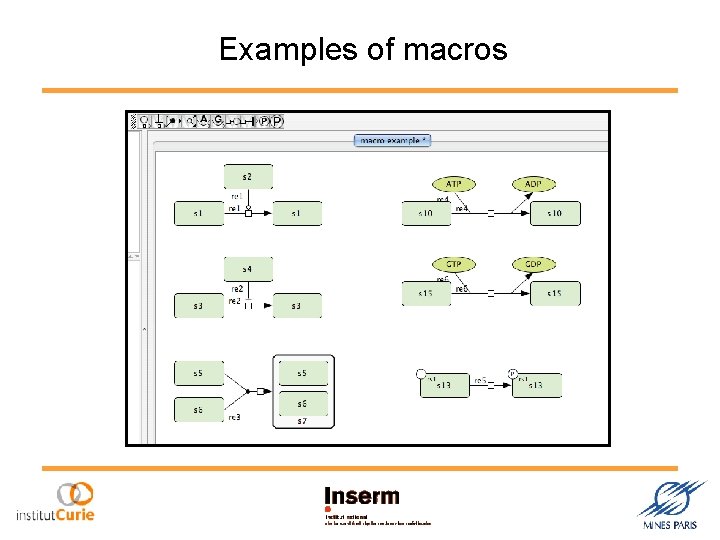
Examples of macros

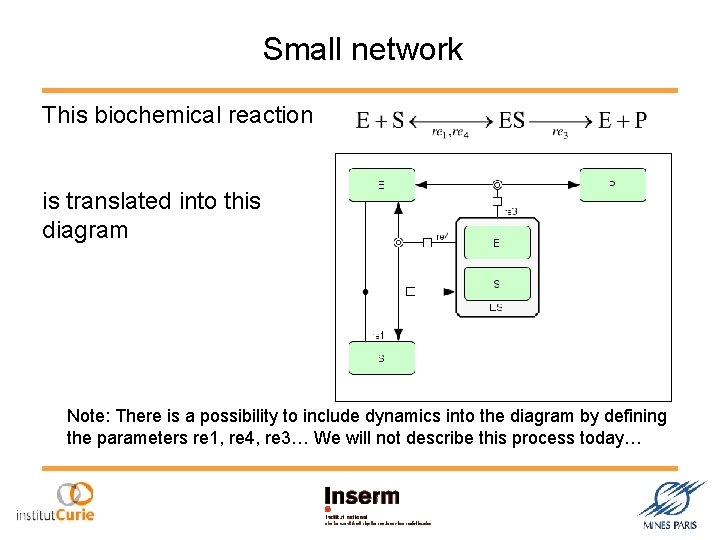
Small network This biochemical reaction is translated into this diagram Note: There is a possibility to include dynamics into the diagram by defining the parameters re 1, re 4, re 3… We will not describe this process today…

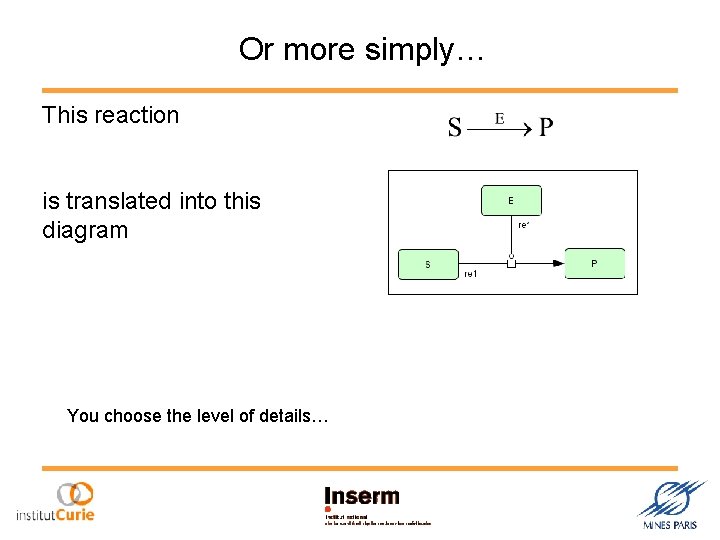
Or more simply… This reaction is translated into this diagram You choose the level of details…
![Some functions Drawing tips Choose from the Edit menu Grid Snap to help Some functions Drawing tips: - Choose from the [Edit] menu, Grid Snap, to help](https://slidetodoc.com/presentation_image_h/dfd16326367e4209811037030ff38f52/image-27.jpg)
Some functions Drawing tips: - Choose from the [Edit] menu, Grid Snap, to help you draw your network. - You can also make the grid visible Grid Snap On or OFF indicated here

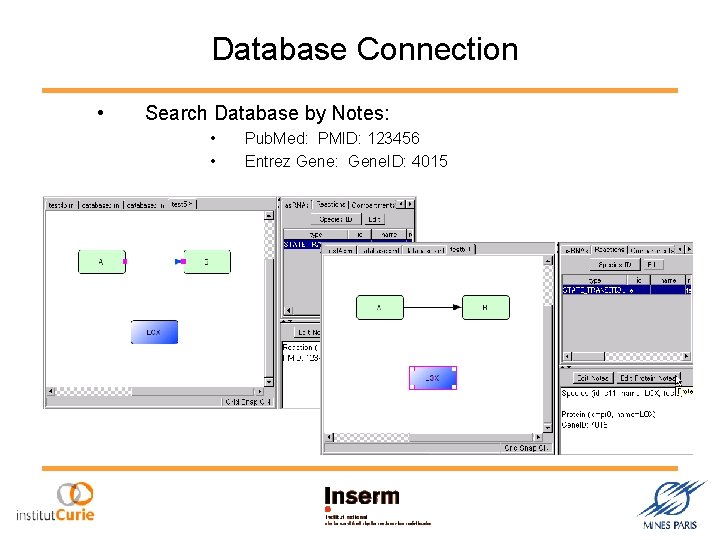
Database Connection • Search Database by Notes: • • Pub. Med: PMID: 123456 Entrez Gene: Gene. ID: 4015

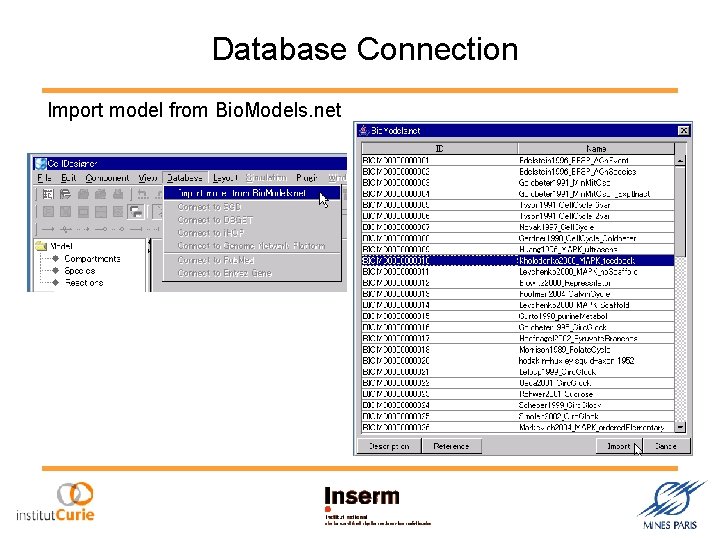
Database Connection Import model from Bio. Models. net

Practice

Inventory of features • • • • Open a new model Create a reaction Add anchor point Add catalytic reaction Set active state Change color Include compartment Add residue to protein Change position of residue Create complex Include genes and RNAs Connect to databases Choose layout

Demo

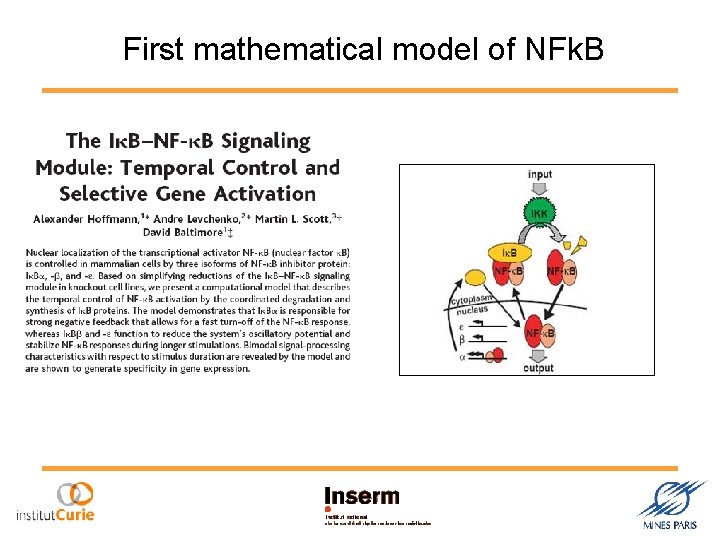
First mathematical model of NFk. B
