Validation of Screening Methods Aisling Treacy MSc Prof


- Slides: 24

Validation of Screening Methods Aisling Treacy, MSc. Prof. Tom Buckley, MSc, FIBMS, FAMLS

Background n n n n Established in 1984 – Equine diagnostic laboratory Screening laboratory during the “Angel Dust Era” Antibiotic residue testing for the porcine industry where there was approx. 25% non-compliance → currently < 1% non-compliance Screening laboratory for the NRCP → obtained ISO 17025 Accreditation for NRCP tests in 2005 → Currently accredited for 94 analyte / matrix / species combinations Rapid screening laboratory for Phenylbutazone (PBZ) during the horse meat scandal – 2013 Self Monitoring Plan Equine anti-doping testing → testing other species

Pre-validation criteria to be considered n Analyte(s) – regulatory limit – MRL, MRPL, RPA n Selection of the method – LOD, cross-reactivity, single / multianalyte method, reliability, matrix n Client requirements n Review of relevant regulatory or guidance documents q q Commission Decision EC/2002/657 CRL Guidance Paper of 7 December 2007 Community Reference Laboratories Residues (CRLs) 20/1/2010 ISO/IEC 17025: 2005 General requirements for the competence of testing and calibration laboratories

Pre-validation work n Test performance – recovery of spikes (various levels), reproducible results n Matrix / species effects n Sample treatment / optimum extraction

Study design / Validation plan n n n n Analyte, matrix and species to be validated Initial validation CCβ, Screening Target Concentration → validation spiking level Number of samples to be analysed Factors to be included in ruggedness Specificity testing with related analytes Applicability Further validation Stability testing Repeatability / method verification

Performance characteristics to be determined according to EC/2002/657 n Semi-quantitative screening: q q q CCβ Precision Selectivity / specificity Applicability Ruggedness Stability

CCβ Detection capability n CCβ: q q q n Detection capability CCβ – is defined as the smallest content of the analyte that may be detected, identified and/or quantified in a sample with an error probability of β The β error is the probability that the tested sample is truly non-compliant even though a compliant measurement has been obtained For screening tests the β error (i. e. false compliant rate) should be <5% For analytes with a regulatory limit, CCβ must be ≤ the regulatory limit For analytes with no regulatory limit, CCβ must be as low as possible CCβ must be established for analytes across individual matrices LOD: q q A consideration when selecting a method CCβ and threshold values to describe the detection capability of the method

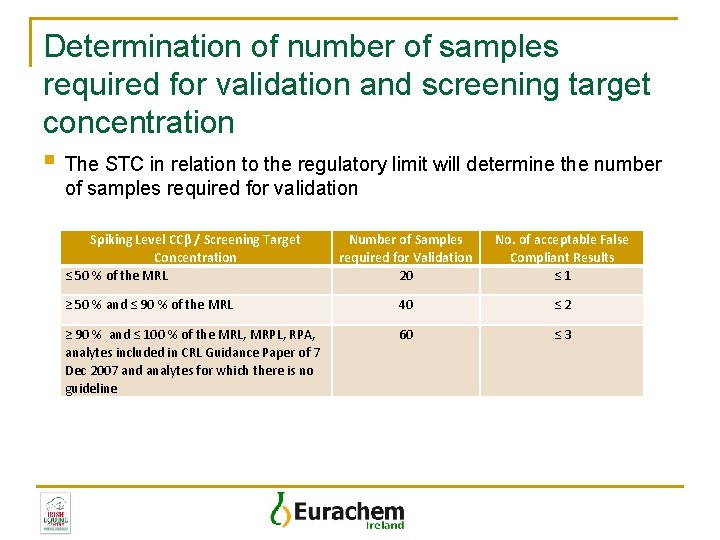
Determination of number of samples required for validation and screening target concentration § The STC in relation to the regulatory limit will determine the number of samples required for validation Spiking Level CCβ / Screening Target Concentration ≤ 50 % of the MRL Number of Samples required for Validation 20 No. of acceptable False Compliant Results ≤ 1 ≥ 50 % and ≤ 90 % of the MRL 40 ≤ 2 ≥ 90 % and ≤ 100 % of the MRL, MRPL, RPA, analytes included in CRL Guidance Paper of 7 Dec 2007 and analytes for which there is no guideline 60 ≤ 3

Implementation of validation n Determination of STC and number of samples required for validation n Analyses carried out on 3 different days by different analysts

Inclusion of other species and matrices / applicability n Separate validation is carried out for each individual matrix type, e. g. serum and muscle n If multiple species are to be validated within one matrix type, then the 60 samples could comprise different species, e. g. 20 x bovine, 20 x porcine and 20 x ovine n If an additional species is to be included after validation is complete, 20 spiked and 20 blank samples made up of the additional species may be analysed and added to the initial validation for the calculation of threshold value and cut-off value

Test specificity n Testing of blank samples to determine threshold will demonstrate whethere is matrix interference – calculation of threshold values n Testing of similar analytes – recovery should fall below cut-off value established during validation

Calculation of cut-off value n Two approaches to establishing cut-off levels outlined in CRL Guidelines for the Validation of Screening Methods for Residues of Veterinary Medicines 2010: q q Approach I – Lowest response The lowest response in the spiked samples analysed during validation is taken as the cut-off giving a response greater than this level is deemed to be ‘screen positive’ and exceeds the CCβ of the screening method Approach II – Statistical approach Cut-off factor (FM) = Mean Response – 1. 64 x SD

Calculation of cut-off value n The multiplication factor approach to establish a cut-off value q q q The IEC uses a multiplication factor applied to the mean of two spiked ‘cut-off’ samples analysed alongside routine samples to establish a cut-off value for each test run A one-tailed 95% confidence interval of the validation data generated from spiked samples is calculated and then divided by the mean (M) To apply this fraction to the results of routine spiked cut-off samples, this figure is subtracted from 1 to give a multiplication factor to be applied to the mean of the spiked cut-off samples Multiplication factor = 1 - (1. 64 x Std. Dev. ) M q In routine testing, any sample with a reading at or above the calculated ‘cut-off applied’ will be considered ‘screen positive’ or non-compliant and forwarded for confirmatory analysis q This approach to cut-off calculation has been approved by regulatory authorities and INAB auditors q

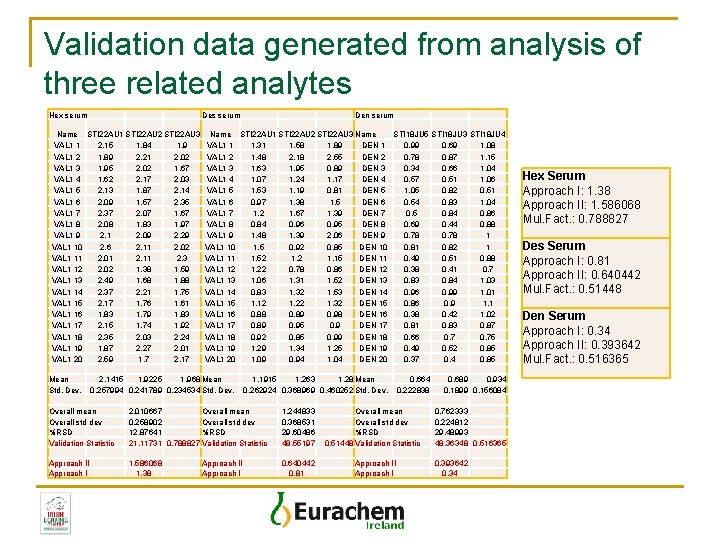
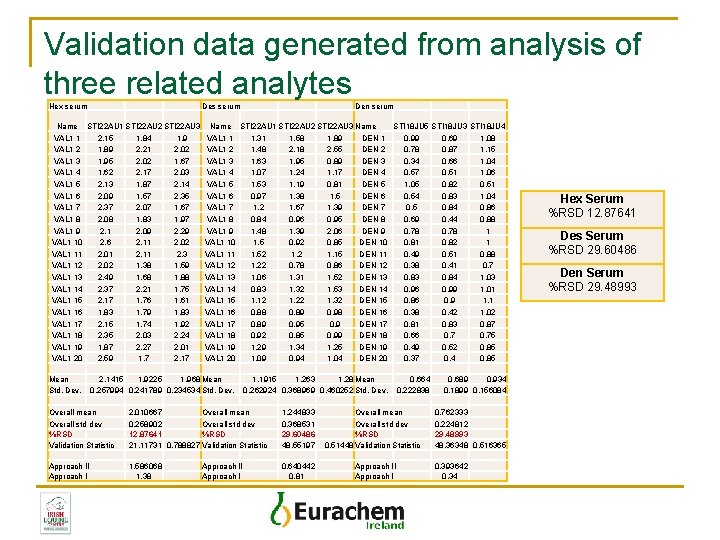
Validation data generated from analysis of three related analytes Hex serum Des serum Name STI 22 AU 1 STI 22 AU 2 STI 22 AU 3 VAL 1 1 2. 15 1. 84 1. 9 VAL 1 2 1. 89 2. 21 2. 02 VAL 1 3 1. 95 2. 02 1. 67 VAL 1 4 1. 62 2. 17 2. 03 VAL 1 5 2. 13 1. 87 2. 14 VAL 1 6 2. 09 1. 57 2. 35 VAL 1 7 2. 37 2. 07 1. 67 VAL 1 8 2. 08 1. 83 1. 97 VAL 1 9 2. 1 2. 09 2. 29 VAL 1 10 2. 6 2. 11 2. 02 VAL 1 11 2. 01 2. 11 2. 3 VAL 1 12 2. 02 1. 38 1. 59 VAL 1 13 2. 49 1. 68 1. 88 VAL 1 14 2. 37 2. 21 1. 75 VAL 1 15 2. 17 1. 76 1. 61 VAL 1 16 1. 83 1. 79 1. 83 VAL 1 17 2. 15 1. 74 1. 92 VAL 1 18 2. 35 2. 03 2. 24 VAL 1 19 1. 87 2. 27 2. 01 VAL 1 20 2. 59 1. 7 2. 17 Mean Std. Dev. Den serum Name STI 22 AU 1 STI 22 AU 2 STI 22 AU 3 Name STI 18 JU 5 STI 18 JU 3 STI 18 JU 4 VAL 1 1 1. 31 1. 58 1. 89 DEN 1 0. 99 0. 69 1. 08 VAL 1 2 1. 48 2. 18 2. 55 DEN 2 0. 78 0. 87 1. 15 VAL 1 3 1. 63 1. 95 0. 89 DEN 3 0. 34 0. 66 1. 04 VAL 1 4 1. 07 1. 24 1. 17 DEN 4 0. 57 0. 51 1. 06 VAL 1 5 1. 53 1. 19 0. 81 DEN 5 1. 05 0. 82 0. 51 VAL 1 6 0. 97 1. 38 1. 5 DEN 6 0. 54 0. 83 1. 04 VAL 1 7 1. 2 1. 67 1. 39 DEN 7 0. 5 0. 84 0. 86 VAL 1 8 0. 84 0. 96 0. 95 DEN 8 0. 69 0. 44 0. 88 VAL 1 9 1. 48 1. 39 2. 06 DEN 9 0. 78 1 VAL 1 10 1. 5 0. 92 0. 85 DEN 10 0. 81 0. 82 1 VAL 1 11 1. 52 1. 15 DEN 11 0. 49 0. 51 0. 88 VAL 1 12 1. 22 0. 78 0. 86 DEN 12 0. 38 0. 41 0. 7 VAL 1 13 1. 06 1. 31 1. 52 DEN 13 0. 84 1. 03 VAL 1 14 0. 83 1. 32 1. 53 DEN 14 0. 96 0. 99 1. 01 VAL 1 15 1. 12 1. 22 1. 32 DEN 15 0. 86 0. 9 1. 1 VAL 1 16 0. 88 0. 89 0. 98 DEN 16 0. 38 0. 42 1. 02 VAL 1 17 0. 89 0. 95 0. 9 DEN 17 0. 81 0. 83 0. 87 VAL 1 18 0. 92 0. 85 0. 99 DEN 18 0. 66 0. 75 VAL 1 19 1. 29 1. 34 1. 25 DEN 19 0. 49 0. 52 0. 85 VAL 1 20 1. 09 0. 94 1. 04 DEN 20 0. 37 0. 4 0. 85 2. 1415 1. 9225 1. 968 Mean 0. 257994 0. 241789 0. 234534 Std. Dev. 1. 1915 1. 263 1. 28 Mean 0. 262924 0. 368969 0. 460252 Std. Dev. Overall mean Overall std dev %RSD Validation Statistic 2. 010667 Overall mean 0. 258902 Overall std dev 12. 87641 %RSD 21. 11731 0. 788827 Validation Statistic 1. 244833 0. 368531 29. 60486 48. 55197 Approach II Approach I 1. 586068 1. 38 0. 640442 0. 81 Approach II Approach I 0. 664 0. 222838 Overall mean Overall std dev %RSD 0. 51448 Validation Statistic Approach II Approach I 0. 689 0. 934 0. 1899 0. 156084 0. 762333 0. 224812 29. 48993 48. 36348 0. 516365 0. 393642 0. 34 Hex Serum Approach I: 1. 38 Approach II: 1. 586068 Mul. Fact. : 0. 788827 Des Serum Approach I: 0. 81 Approach II: 0. 640442 Mul. Fact. : 0. 51448 Den Serum Approach I: 0. 34 Approach II: 0. 393642 Mul. Fact. : 0. 516365

Ruggedness n Introduction of minor reasonable variations and observations of consequences n The Youden approach to ruggedness is employed at the IEC – facilitates the introduction of several variations simultaneously n Factors that may influence measurement result are selected: q q q n Pre-treatment Clean-up, SPE Analysis Factors that influence results are subjected to further testing – these factors are described in the validation report and may be included in the SOP

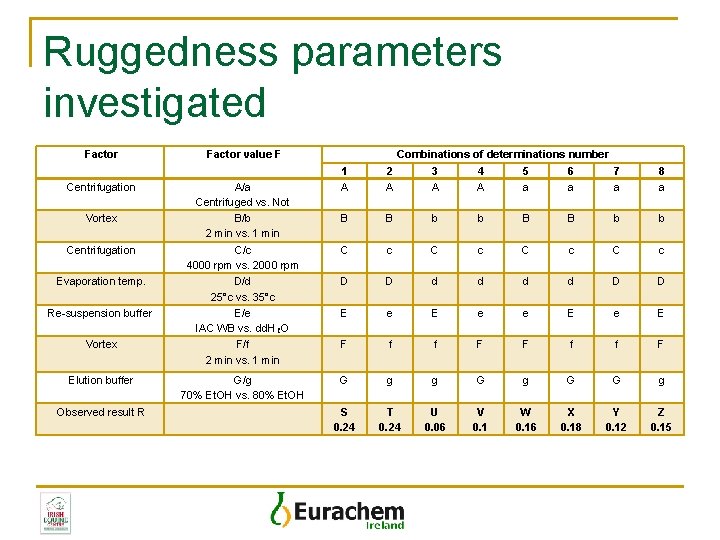
Ruggedness parameters investigated Factor Centrifugation Vortex Centrifugation Evaporation temp. Re-suspension buffer Vortex Elution buffer Observed result R Factor value F Combinations of determinations number 1 A 2 A 3 A 4 A 5 a 6 a 7 a 8 a B B b b C/c 4000 rpm vs. 2000 rpm D/d 25°c vs. 35°c E/e IAC WB vs. dd. H₂O F/f 2 min vs. 1 min C c C c D D d d D D E e e E F f f F G/g 70% Et. OH vs. 80% Et. OH G g g G G g S 0. 24 T 0. 24 U 0. 06 V 0. 1 W 0. 16 X 0. 18 Y 0. 12 Z 0. 15 A/a Centrifuged vs. Not B/b 2 min vs. 1 min

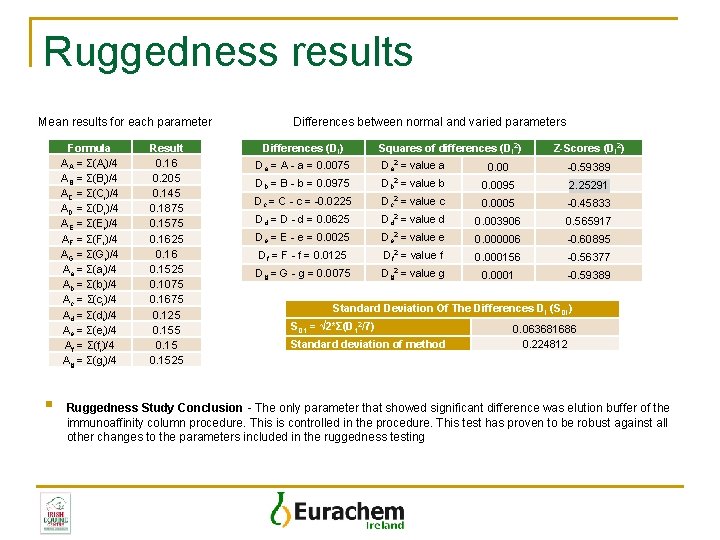
Ruggedness results Mean results for each parameter Formula AA = Σ(Ai)/4 AB = Σ(Bi)/4 AC = Σ(Ci)/4 AD = Σ(Di)/4 AE = Σ(Ei)/4 AF = Σ(Fi)/4 AG = Σ(Gi)/4 Aa = Σ(ai)/4 Ab = Σ(bi)/4 Ac = Σ(ci)/4 Ad = Σ(di)/4 Ae = Σ(ei)/4 Af = Σ(fi)/4 Ag = Σ(gi)/4 § Result 0. 16 0. 205 0. 145 0. 1875 0. 1575 0. 1625 0. 16 0. 1525 0. 1075 0. 1675 0. 125 0. 1525 Differences between normal and varied parameters Differences (Di) Squares of differences (Di 2) Z-Scores (Di 2) Da = A - a = 0. 0075 Da 2 = value a 0. 00 -0. 59389 Db = B - b = 0. 0975 Db 2 = value b 0. 0095 2. 25291 Dc = C - c = -0. 0225 Dc 2 = value c 0. 0005 -0. 45833 Dd = D - d = 0. 0625 Dd 2 = value d 0. 003906 0. 565917 De = E - e = 0. 0025 De 2 = value e 0. 000006 -0. 60895 Df = F - f = 0. 0125 Df 2 = value f 0. 000156 -0. 56377 Dg = G - g = 0. 0075 Dg 2 = value g 0. 0001 -0. 59389 Standard Deviation Of The Differences DI (SDI) SD 1 = √ 2*Σ(D 12/7) Standard deviation of method 0. 063681686 0. 224812 Ruggedness Study Conclusion - The only parameter that showed significant difference was elution buffer of the immunoaffinity column procedure. This is controlled in the procedure. This test has proven to be robust against all other changes to the parameters included in the ruggedness testing

Stability n The degradation of the analyte under different conditions relevant to the test / laboratory n Observed through ongoing monitoring of QC materials (negatives and spikes) – plotted on Shewhart charts n Stability experiments carried out if any degradation is observed over time n Stability experiments – assessment of stability of analytes: q q q Stability of analytes in solution Stability of analytes in matrix Assessment carried out in line with EC/2002/657 and SOP P 5. 5. 135

Repeatability n Assessment of spiked samples over three days n Calculation of Relative Standard Deviation measure of within lab reproducibility must be < 35%

Validation acceptability criteria – Was validation successful? n RSD < 35% n Approach I – No overlap between threshold value and cut-off value n Approach II – The rate of false positive is acceptable (i. e. 5%) when FM > T if B < FM < T the false positive rate is higher than 5% n According to Commission Decision 2002/657/EC, CCβ is validated when FM > B n If the above criteria are met the method is considered robust, specific and fit for purpose

Validation data generated from analysis of three related analytes Hex serum Des serum Name STI 22 AU 1 STI 22 AU 2 STI 22 AU 3 VAL 1 1 2. 15 1. 84 1. 9 VAL 1 2 1. 89 2. 21 2. 02 VAL 1 3 1. 95 2. 02 1. 67 VAL 1 4 1. 62 2. 17 2. 03 VAL 1 5 2. 13 1. 87 2. 14 VAL 1 6 2. 09 1. 57 2. 35 VAL 1 7 2. 37 2. 07 1. 67 VAL 1 8 2. 08 1. 83 1. 97 VAL 1 9 2. 1 2. 09 2. 29 VAL 1 10 2. 6 2. 11 2. 02 VAL 1 11 2. 01 2. 11 2. 3 VAL 1 12 2. 02 1. 38 1. 59 VAL 1 13 2. 49 1. 68 1. 88 VAL 1 14 2. 37 2. 21 1. 75 VAL 1 15 2. 17 1. 76 1. 61 VAL 1 16 1. 83 1. 79 1. 83 VAL 1 17 2. 15 1. 74 1. 92 VAL 1 18 2. 35 2. 03 2. 24 VAL 1 19 1. 87 2. 27 2. 01 VAL 1 20 2. 59 1. 7 2. 17 Mean Std. Dev. Den serum Name STI 22 AU 1 STI 22 AU 2 STI 22 AU 3 Name STI 18 JU 5 STI 18 JU 3 STI 18 JU 4 VAL 1 1 1. 31 1. 58 1. 89 DEN 1 0. 99 0. 69 1. 08 VAL 1 2 1. 48 2. 18 2. 55 DEN 2 0. 78 0. 87 1. 15 VAL 1 3 1. 63 1. 95 0. 89 DEN 3 0. 34 0. 66 1. 04 VAL 1 4 1. 07 1. 24 1. 17 DEN 4 0. 57 0. 51 1. 06 VAL 1 5 1. 53 1. 19 0. 81 DEN 5 1. 05 0. 82 0. 51 VAL 1 6 0. 97 1. 38 1. 5 DEN 6 0. 54 0. 83 1. 04 VAL 1 7 1. 2 1. 67 1. 39 DEN 7 0. 5 0. 84 0. 86 VAL 1 8 0. 84 0. 96 0. 95 DEN 8 0. 69 0. 44 0. 88 VAL 1 9 1. 48 1. 39 2. 06 DEN 9 0. 78 1 VAL 1 10 1. 5 0. 92 0. 85 DEN 10 0. 81 0. 82 1 VAL 1 11 1. 52 1. 15 DEN 11 0. 49 0. 51 0. 88 VAL 1 12 1. 22 0. 78 0. 86 DEN 12 0. 38 0. 41 0. 7 VAL 1 13 1. 06 1. 31 1. 52 DEN 13 0. 84 1. 03 VAL 1 14 0. 83 1. 32 1. 53 DEN 14 0. 96 0. 99 1. 01 VAL 1 15 1. 12 1. 22 1. 32 DEN 15 0. 86 0. 9 1. 1 VAL 1 16 0. 88 0. 89 0. 98 DEN 16 0. 38 0. 42 1. 02 VAL 1 17 0. 89 0. 95 0. 9 DEN 17 0. 81 0. 83 0. 87 VAL 1 18 0. 92 0. 85 0. 99 DEN 18 0. 66 0. 75 VAL 1 19 1. 29 1. 34 1. 25 DEN 19 0. 49 0. 52 0. 85 VAL 1 20 1. 09 0. 94 1. 04 DEN 20 0. 37 0. 4 0. 85 2. 1415 1. 9225 1. 968 Mean 0. 257994 0. 241789 0. 234534 Std. Dev. 1. 1915 1. 263 1. 28 Mean 0. 262924 0. 368969 0. 460252 Std. Dev. Overall mean Overall std dev %RSD Validation Statistic 2. 010667 Overall mean 0. 258902 Overall std dev 12. 87641 %RSD 21. 11731 0. 788827 Validation Statistic 1. 244833 0. 368531 29. 60486 48. 55197 Approach II Approach I 1. 586068 1. 38 0. 640442 0. 81 Approach II Approach I 0. 664 0. 222838 Overall mean Overall std dev %RSD 0. 51448 Validation Statistic Approach II Approach I 0. 689 0. 934 0. 1899 0. 156084 0. 762333 0. 224812 29. 48993 48. 36348 0. 516365 0. 393642 0. 34 Hex Serum %RSD 12. 87641 Des Serum %RSD 29. 60486 Den Serum %RSD 29. 48993

When is additional or re-validation required? n Additional validation carried out when extra species are introduced or minor changes to an existing method n Re-validation will occur if: q q q Significant change to the method Method fails on a continual basis Change in client requirements, e. g. lower STC

Continuous verification n QC samples q q q Negative control (TQCN) Positive controls (Cut-off 1 and 2) Check sample (blind to analyst) n Spiking analyte – the analyte with the poorest recovery (worst case / lowest specificity) is used to spike QC or most relevant analyte according to test n Last 20 sets of QC analyses plotted on Shewhart charts - ≤ 1 outlier per 20 analyses, i. e. ≤ 5% q n PT’s – CRL / State Lab submit samples for PT analysis for each analyte at least once yearly q q n Verifies overall method performance, analyst accuracy / performance, spike stability, kit performance / stability External providers – Progetto Trieste, FAPAS In-house Out of specification results subjected to an in-house investigation procedure

Thank You