Requirements for oxidative phosphorylation Requirements for the production






- Slides: 29

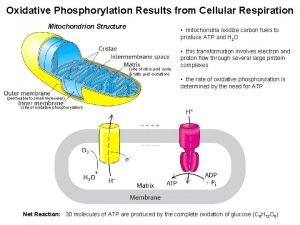
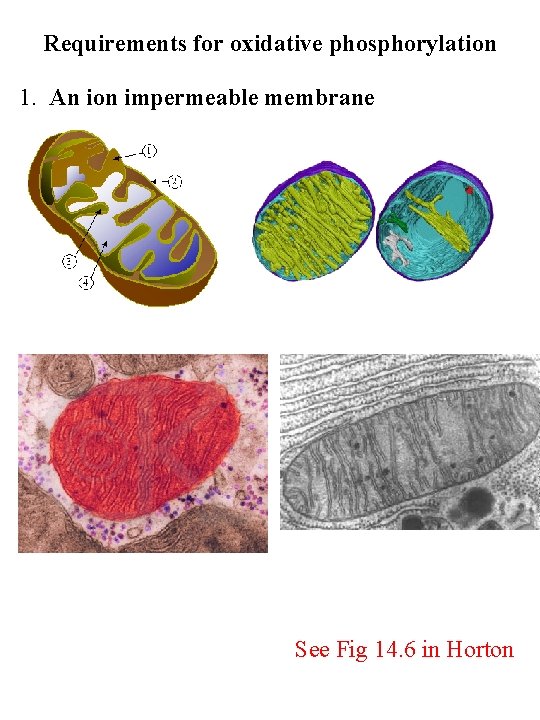
Requirements for oxidative phosphorylation Requirements for the production of ATP 1. An ion impermeable membrane 2. A mechanism for moving protons (H+) across the membrane to produce an energy-rich proton gradient 3. A mechanism to capture the energy made available as protons move down the proton gradient

Requirements for oxidative phosphorylation 1. An ion impermeable membrane See Fig 14. 6 in Horton

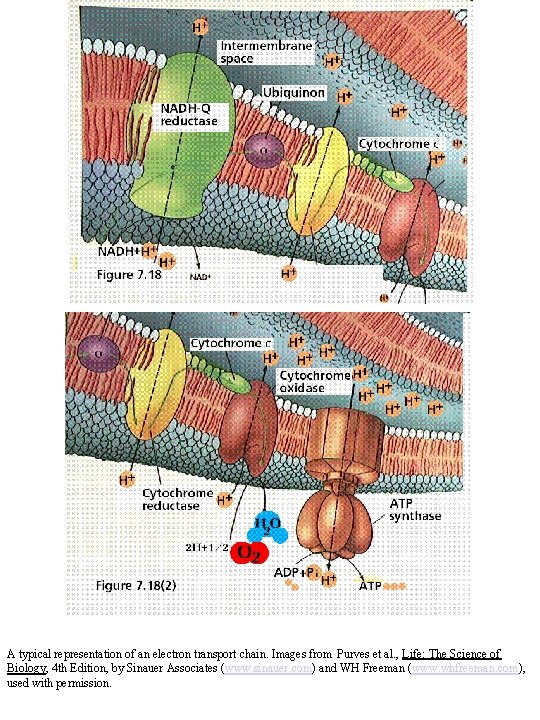
A typical representation of an electron transport chain. Images from Purves et al. , Life: The Science of Biology, 4 th Edition, by Sinauer Associates (www. sinauer. com) and WH Freeman (www. whfreeman. com), used with permission.

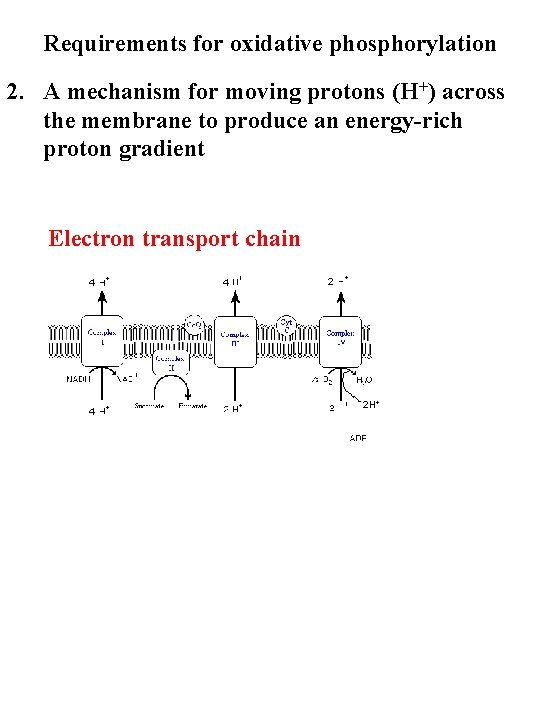
Requirements for oxidative phosphorylation 2. A mechanism for moving protons (H+) across the membrane to produce an energy-rich proton gradient Electron transport chain 4 4 4 2 2 H+

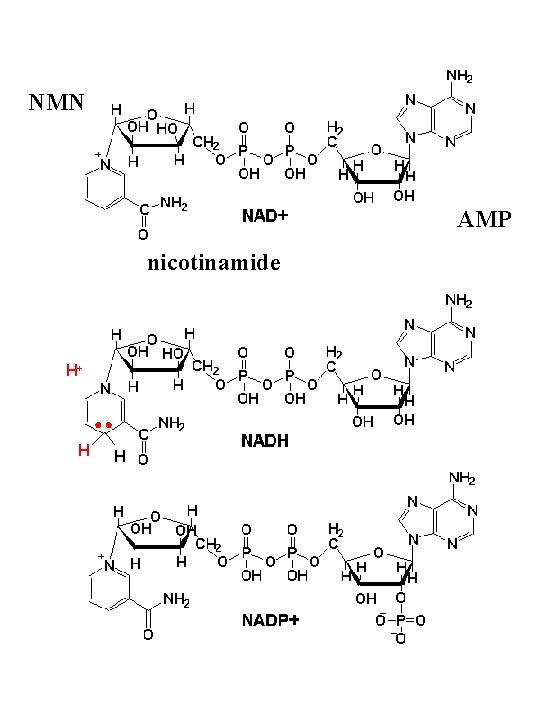
NMN AMP nicotinamide H+ H H

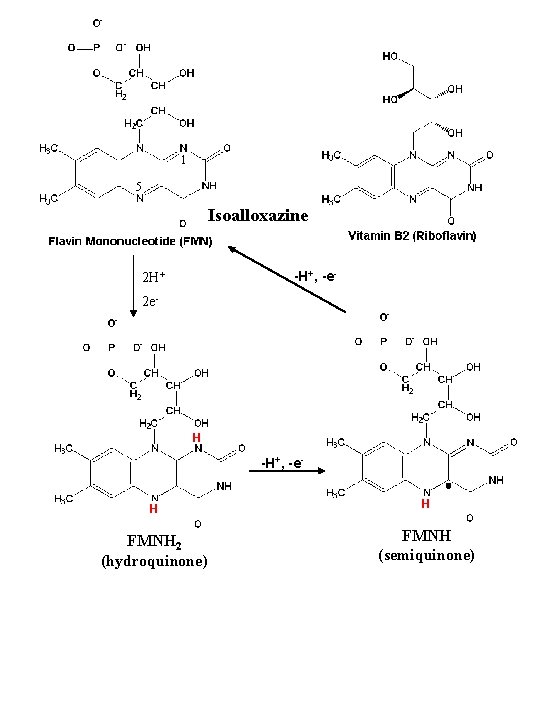
1 5 Isoalloxazine -H+, -e- 2 H+ 2 e- H -H+, -e. H H FMNH 2 (hydroquinone) FMNH (semiquinone)

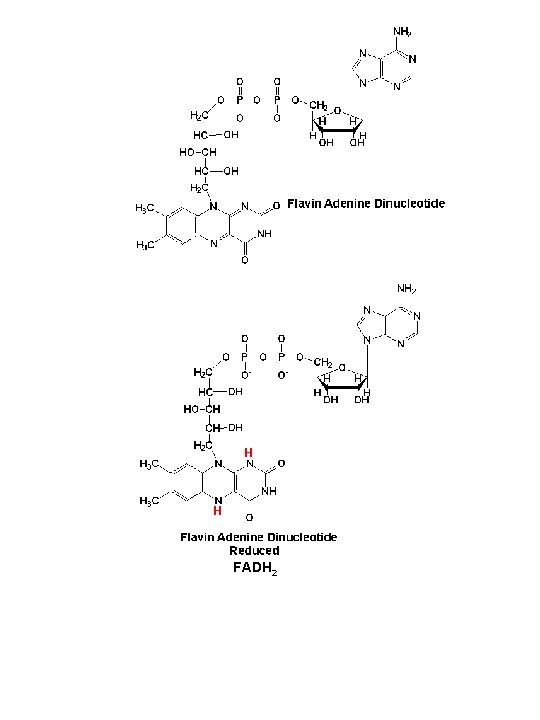
H H FADH 2

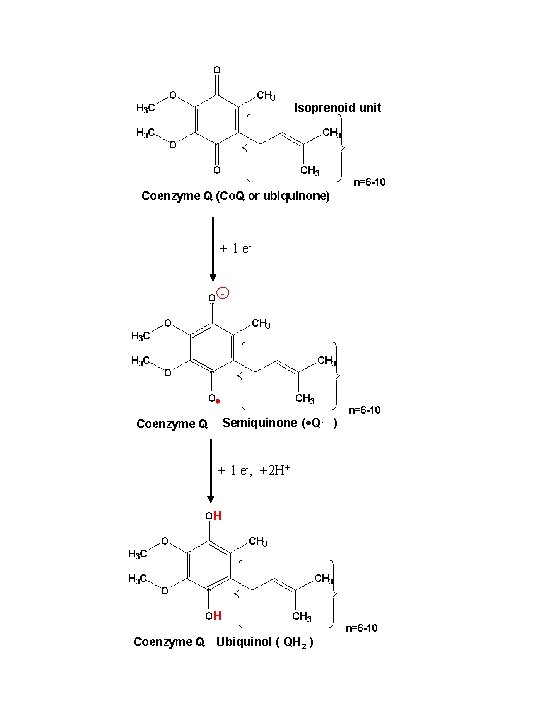
Isoprenoid unit + 1 e- Semiquinone ( Q- ) + 1 e-, +2 H+ H H Ubiquinol ( QH 2 )

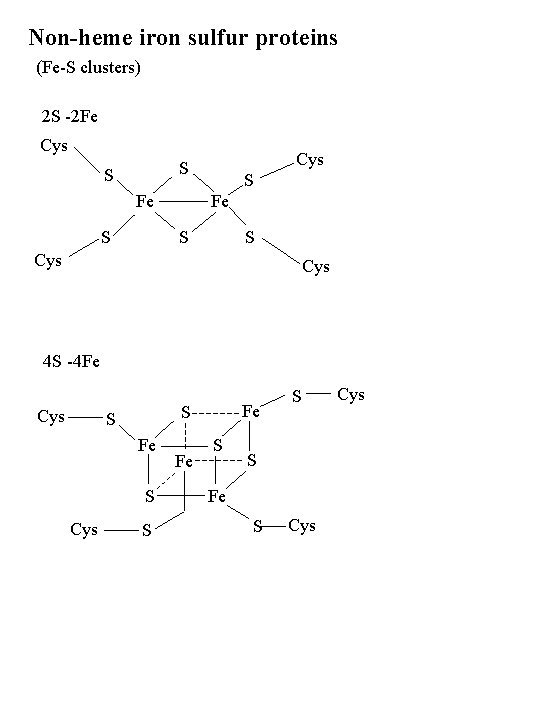
Non-heme iron sulfur proteins (Fe-S clusters) 2 S -2 Fe Cys S S Fe S S Cys 4 S -4 Fe Cys Fe S S S Fe S Cys

Cytochromes • Heme-containing proteins • Classified as a, b or c based on absorption spectrum • Electron transport has: a and a 3, b 566 (b. L) and b 562 (b. H). and c 1 • carry 1 electron per heme iron • a, a 3, b 566, , , b 562, , and c 1 are integral membrane proteins • c is a peripheral membrane protein on outer surface of inner mitochondrial membrane

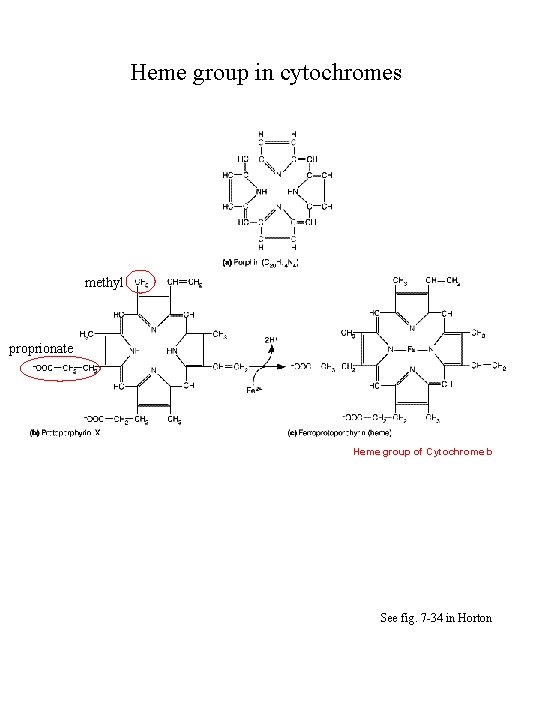
Heme group in cytochromes methyl proprionate Heme group of Cytochrome b See fig. 7 -34 in Horton

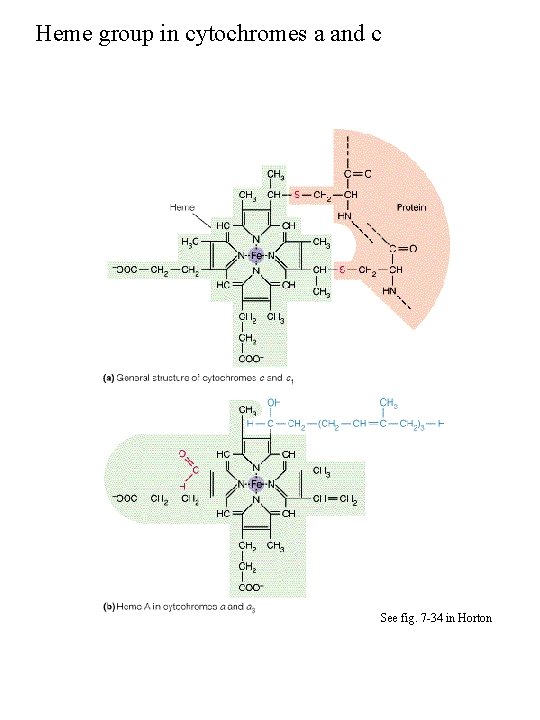
Heme group in cytochromes a and c See fig. 7 -34 in Horton

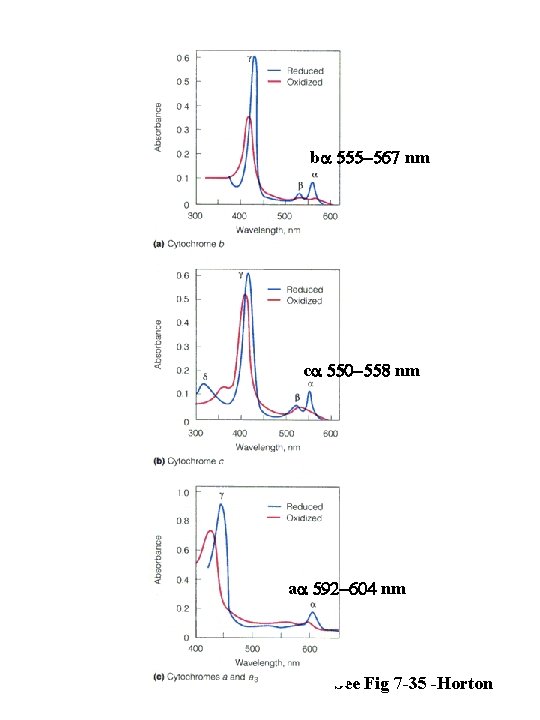
ba 555 -567 nm ca 550 -558 nm aa 592 -604 nm See Fig 7 -35 -Horton

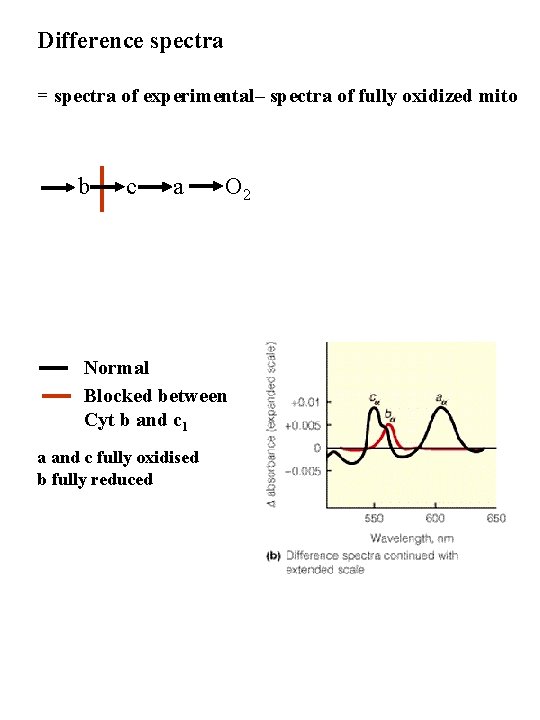
Difference spectra = spectra of experimental– spectra of fully oxidized mito b c a O 2 Normal Blocked between Cyt b and c 1 a and c fully oxidised b fully reduced

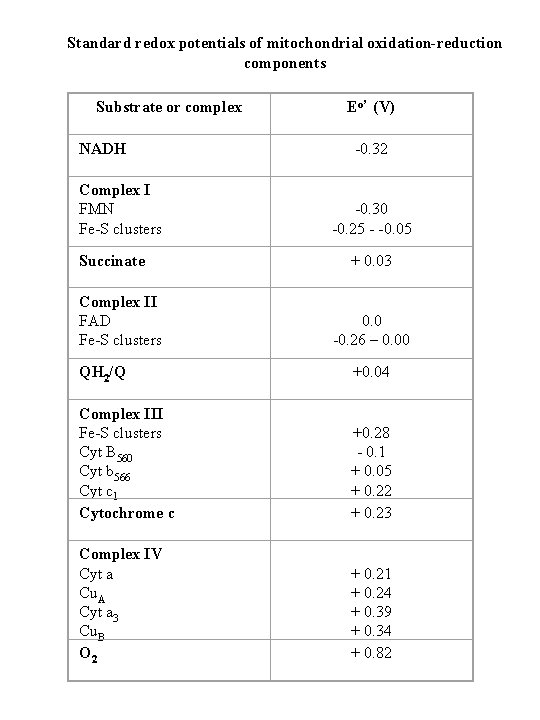
Standard redox potentials of mitochondrial oxidation-reduction components Substrate or complex NADH Complex I FMN Fe-S clusters Succinate Complex II FAD Fe-S clusters Eo’ (V) -0. 32 -0. 30 -0. 25 - -0. 05 + 0. 03 0. 0 -0. 26 – 0. 00 QH 2/Q +0. 04 Complex III Fe-S clusters Cyt B 560 Cyt b 566 Cyt c 1 Cytochrome c +0. 28 - 0. 1 + 0. 05 + 0. 22 + 0. 23 Complex IV Cyt a Cu. A Cyt a 3 Cu. B O 2 + 0. 21 + 0. 24 + 0. 39 + 0. 34 + 0. 82

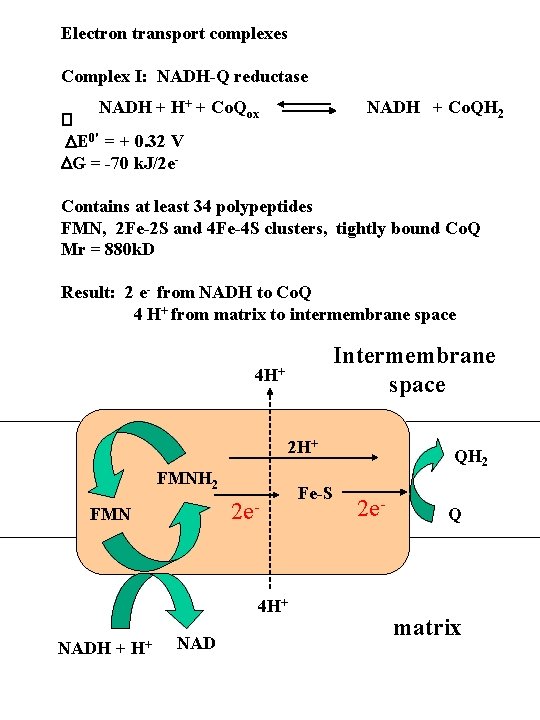
Electron transport complexes Complex I: NADH-Q reductase NADH + H+ + Co. Qox NADH + Co. QH 2 � DE 0' = + 0. 32 V DG = -70 k. J/2 e Contains at least 34 polypeptides FMN, 2 Fe-2 S and 4 Fe-4 S clusters, tightly bound Co. Q Mr = 880 k. D Result: 2 e- from NADH to Co. Q 4 H+ from matrix to intermembrane space Intermembrane space 4 H+ 2 H+ FMNH 2 2 e- FMN 4 H+ NADH + H+ NAD Fe-S QH 2 2 e- Q matrix

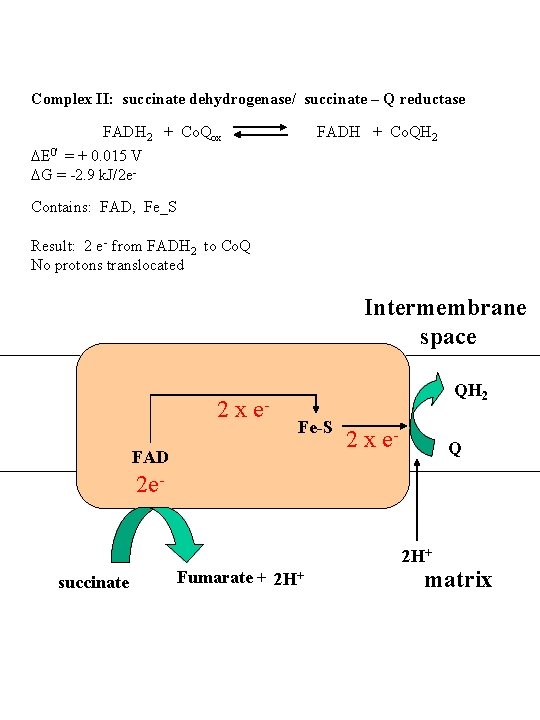
Complex II: succinate dehydrogenase/ succinate – Q reductase FADH 2 + Co. Qox FADH + Co. QH 2 DE 0' = + 0. 015 V DG = -2. 9 k. J/2 e Contains: FAD, Fe_S Result: 2 e- from FADH 2 to Co. Q No protons translocated Intermembrane space 2 x e- QH 2 Fe-S FAD 2 x e- Q 2 e 2 H+ succinate Fumarate + 2 H+ matrix

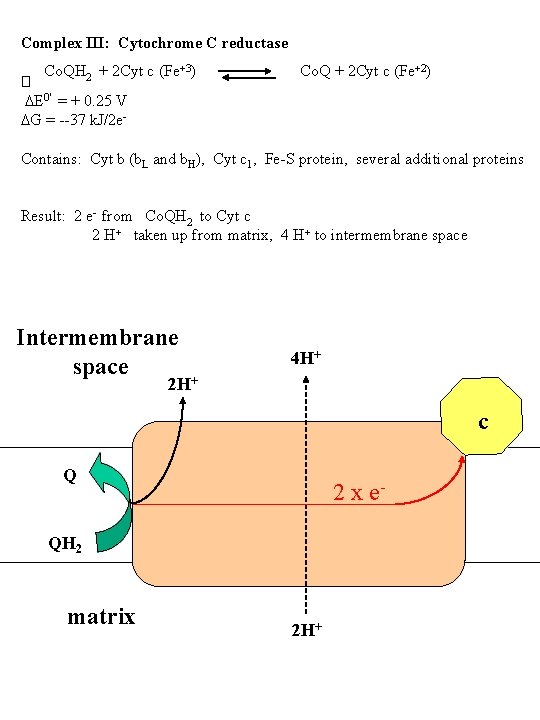
Complex III: Cytochrome C reductase Co. QH 2 + 2 Cyt c (Fe+3) Co. Q + 2 Cyt c (Fe+2) � DE 0' = + 0. 25 V DG = --37 k. J/2 e Contains: Cyt b (b. L and b. H), Cyt c 1, Fe-S protein, several additional proteins Result: 2 e- from Co. QH 2 to Cyt c 2 H+ taken up from matrix, 4 H+ to intermembrane space Intermembrane space + 4 H+ 2 H c Q 2 x e- QH 2 matrix 2 H+

Co. Q cycle C 1 Fe-S b. L b. H C 1 Fe-S b. H b. L

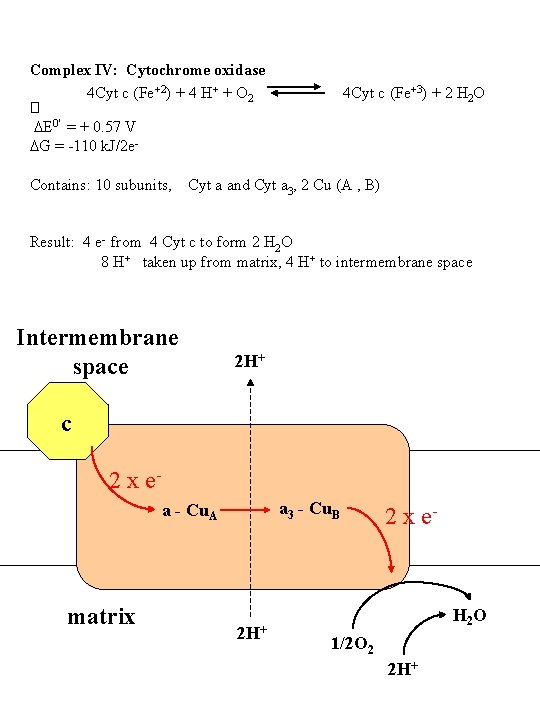
Complex IV: Cytochrome oxidase 4 Cyt c (Fe+2) + 4 H+ + O 2 4 Cyt c (Fe+3) + 2 H 2 O � DE 0' = + 0. 57 V DG = -110 k. J/2 e Contains: 10 subunits, Cyt a and Cyt a 3, 2 Cu (A , B) Result: 4 e- from 4 Cyt c to form 2 H 2 O 8 H+ taken up from matrix, 4 H+ to intermembrane space Intermembrane space 2 H+ c 2 x ea 3 - Cu. B a - Cu. A matrix 2 H+ 2 x e- H 2 O 1/2 O 2 2 H+

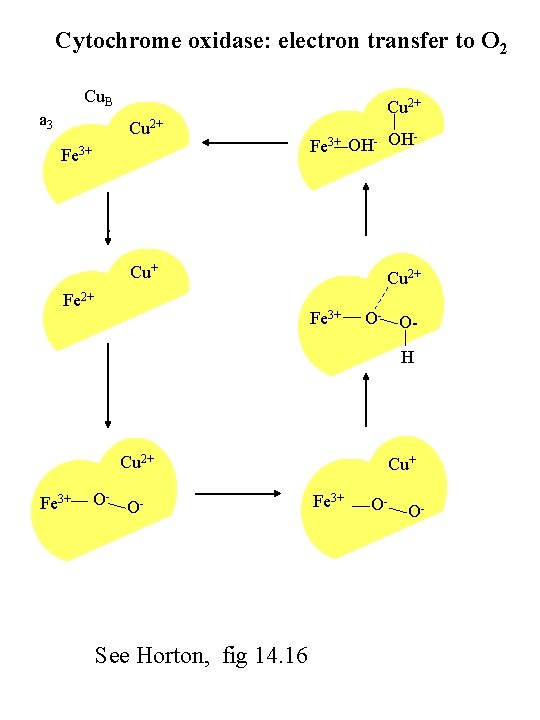
Cytochrome oxidase: electron transfer to O 2 Cu. B a 3 Cu 2+ Fe 3+ OH- OH . Cu+ Fe 2+ Cu 2+ Fe 3+ O- OH Cu 2+ Fe 3+ O- O- See Horton, fig 14. 16 Cu+ Fe 3+ O- O-

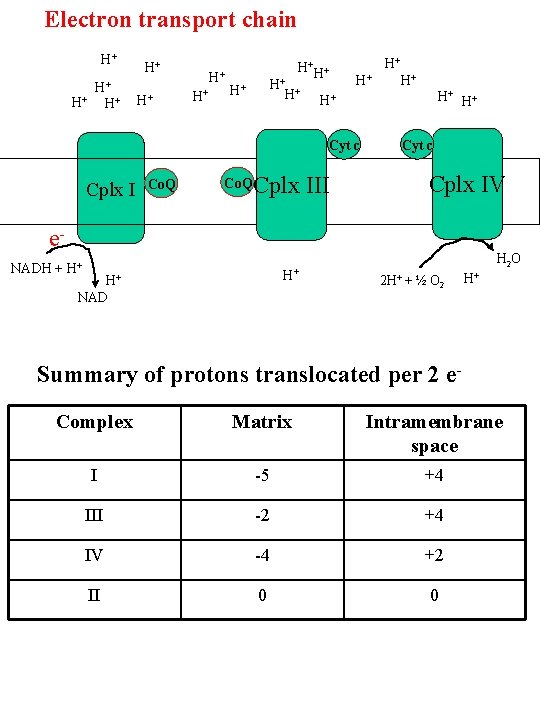
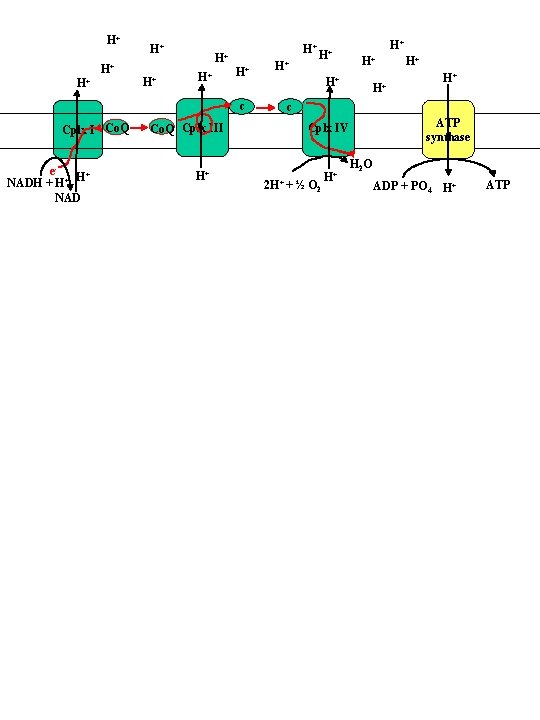
Electron transport chain H+ H+ + H H+ H+ H+ Co. Q H+ H+ H+ Cyt c Cplx I H+ Cplx III Cyt c Cplx IV e. NADH + H+ H+ NAD H 2 O 2 H+ + ½ O 2 H+ Summary of protons translocated per 2 e. Complex Matrix Intramembrane space I -5 +4 III -2 +4 IV -4 +2 II 0 0


1997 Nobel Prize for Chemistry Paul D. Boyer John E. Walker Jens C. Skou "for their elucidation of the enzymatic mechanism underlying the synthesis of adenosine triphosphate (ATP)" "for the first discovery of an ion-transporting enzyme, Na+, K+ -ATPase"

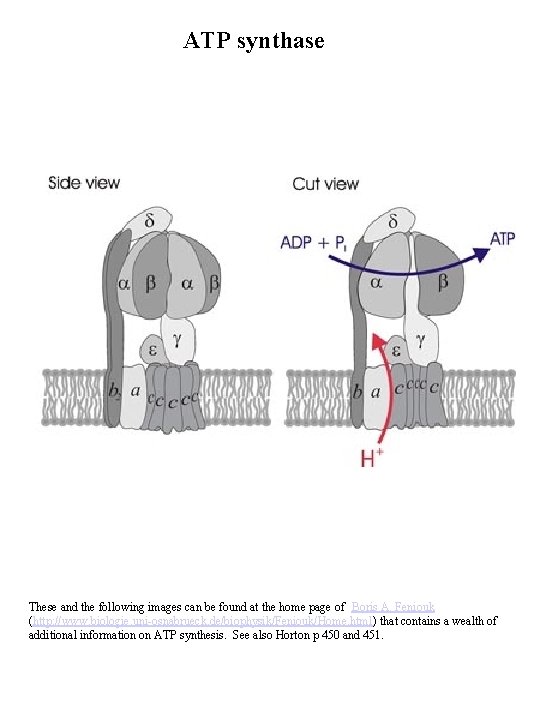
ATP synthase These and the following images can be found at the home page of Boris A. Feniouk (http: //www. biologie. uni-osnabrueck. de/biophysik/Feniouk/Home. html) that contains a wealth of additional information on ATP synthesis. See also Horton p 450 and 451.


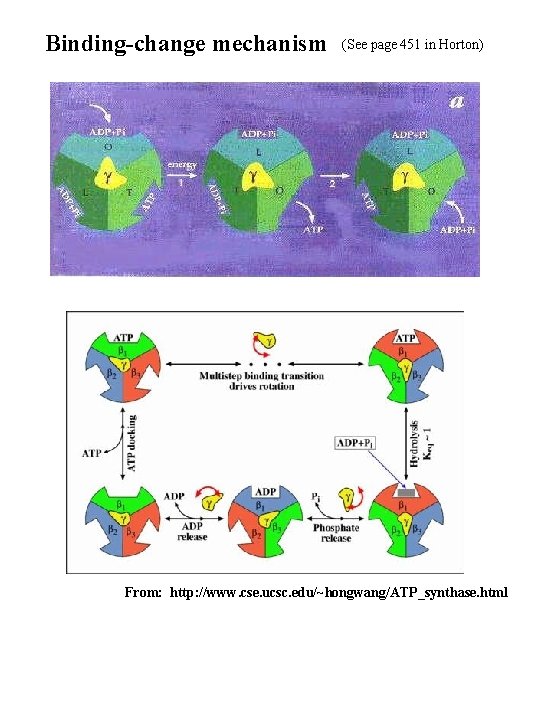
Binding-change mechanism (See page 451 in Horton) From: http: //www. cse. ucsc. edu/~hongwang/ATP_synthase. html

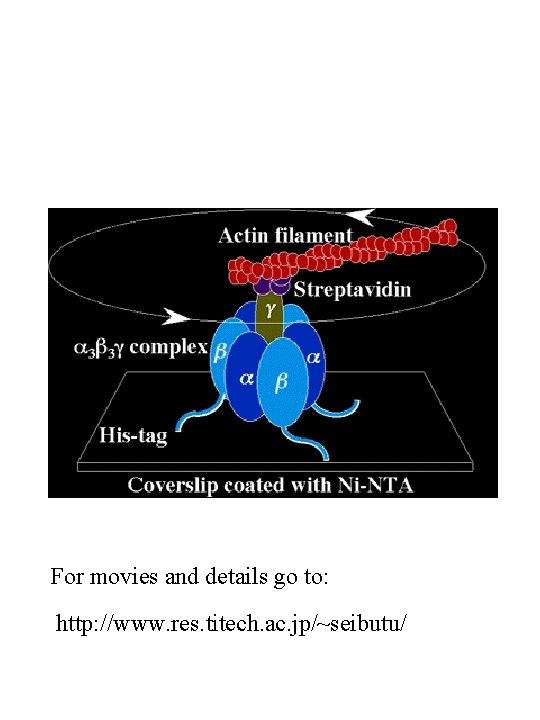
For movies and details go to: http: //www. res. titech. ac. jp/~seibutu/

H+ H+ c Cplx I Co. Q e. H+ NADH + H+ NAD Co. Q Cplx III H+ H+ H+ c ATP synthase Cplx IV 2 H+ + ½ O 2 H+ H 2 O ADP + PO 4 H+ ATP
 Oxidative phosphorylation
Oxidative phosphorylation Uncouplers of oxidative phosphorylation
Uncouplers of oxidative phosphorylation Uncouplers of oxidative phosphorylation
Uncouplers of oxidative phosphorylation Concept map about oxidative phosphorylation
Concept map about oxidative phosphorylation Azide electron transport chain
Azide electron transport chain Phosphorylation definition
Phosphorylation definition Oxidative phosphorylation
Oxidative phosphorylation Oxidative phosphorylation enzymes
Oxidative phosphorylation enzymes Oxidative phosphorylation
Oxidative phosphorylation Inhibitors of oxidative phosphorylation
Inhibitors of oxidative phosphorylation Substrate level phosphorylation vs oxidative
Substrate level phosphorylation vs oxidative Define oxidative phosphorylation
Define oxidative phosphorylation Oxidative phosphorylation energy yield
Oxidative phosphorylation energy yield Uncouple oxidative phosphorylation
Uncouple oxidative phosphorylation Inhibitors of oxidative phosphorylation
Inhibitors of oxidative phosphorylation Uncouplers of oxidative phosphorylation
Uncouplers of oxidative phosphorylation Apa itu pre-production?
Apa itu pre-production? Découplage de la phosphorylation oxydative
Découplage de la phosphorylation oxydative Direct phosphorylation
Direct phosphorylation Copyright
Copyright Phosphorylation cascade
Phosphorylation cascade Substrate level phosphorylation
Substrate level phosphorylation Incomplete tetanus muscle contraction
Incomplete tetanus muscle contraction Sémilogie
Sémilogie Phosphorylation cascade
Phosphorylation cascade Oxidative deamination of amino acids
Oxidative deamination of amino acids Transamination and oxidative deamination
Transamination and oxidative deamination Pentose phosphate pathway diagram
Pentose phosphate pathway diagram Plp mechanism transamination
Plp mechanism transamination Pentose phosphate pathway
Pentose phosphate pathway