Reconstruction on trees and Phylogeny 3 Elchanan Mossel

![Phase transition for the CFN model • Th 1[M 2003]: Suppose that n=3 £ Phase transition for the CFN model • Th 1[M 2003]: Suppose that n=3 £](https://slidetodoc.com/presentation_image_h2/2996d4de6919079becc87df885166260/image-10.jpg)
![CFN Logarithmic reconstruction Th 2[M 2003]: If T is an X-tree on n leaves CFN Logarithmic reconstruction Th 2[M 2003]: If T is an X-tree on n leaves](https://slidetodoc.com/presentation_image_h2/2996d4de6919079becc87df885166260/image-11.jpg)
![Main Lemma [M 2003] • Lemma: Suppose that 2 min 2 > 1, then Main Lemma [M 2003] • Lemma: Suppose that 2 min 2 > 1, then](https://slidetodoc.com/presentation_image_h2/2996d4de6919079becc87df885166260/image-12.jpg)
![Reconstructing the topology [M 2003] • The algorithm: Repeat the following: – Reconstruct the Reconstructing the topology [M 2003] • The algorithm: Repeat the following: – Reconstruct the](https://slidetodoc.com/presentation_image_h2/2996d4de6919079becc87df885166260/image-17.jpg)
![Proving main Lemma • Need to estimate E[σr Maj(σ∂)]. Estimate has two parts: • Proving main Lemma • Need to estimate E[σr Maj(σ∂)]. Estimate has two parts: •](https://slidetodoc.com/presentation_image_h2/2996d4de6919079becc87df885166260/image-18.jpg)
- Slides: 18

Reconstruction on trees and Phylogeny 3 Elchanan Mossel, U. C. Berkeley mossel@stat. berkeley. edu, http: //www. cs. berkeley. edu/~mossel/ Supported by Microsoft Research and the Miller Institute 12/23/2021 1

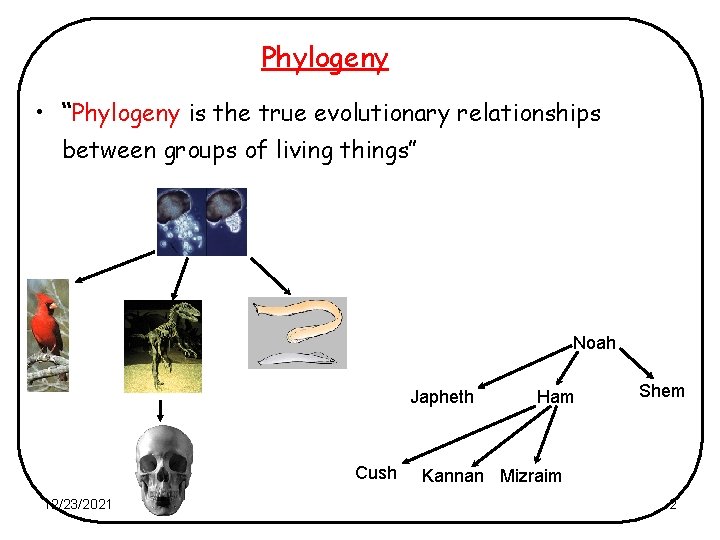
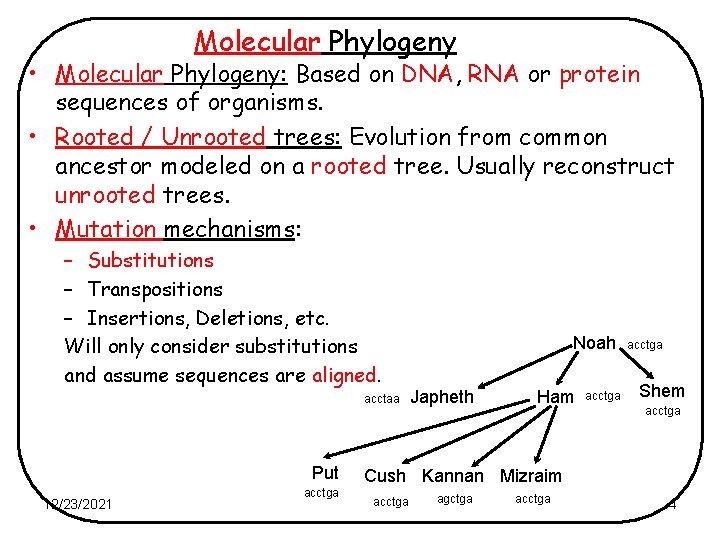
Phylogeny • “Phylogeny is the true evolutionary relationships between groups of living things” Noah Japheth Cush 12/23/2021 Ham Shem Kannan Mizraim 2

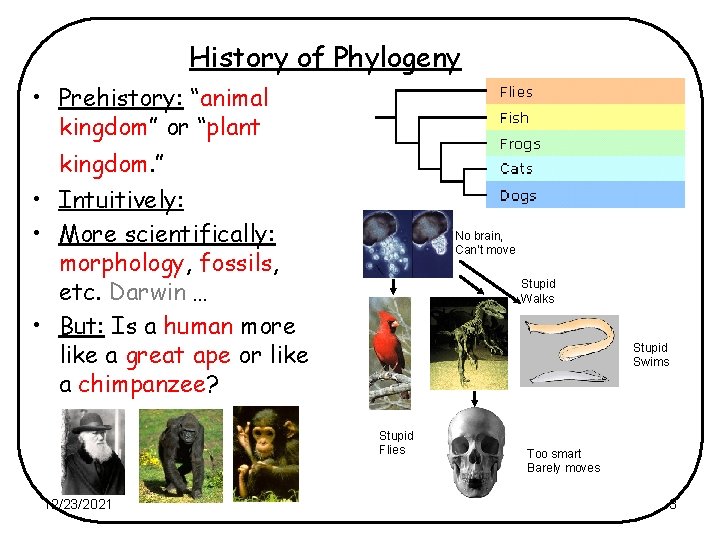
History of Phylogeny • Prehistory: “animal kingdom” or “plant kingdom. ” • Intuitively: • More scientifically: morphology, fossils, etc. Darwin … • But: Is a human more like a great ape or like a chimpanzee? No brain, Can’t move Stupid Walks Stupid Swims Stupid Flies 12/23/2021 Too smart Barely moves 3

Molecular Phylogeny • Molecular Phylogeny: Based on DNA, RNA or protein sequences of organisms. • Rooted / Unrooted trees: Evolution from common ancestor modeled on a rooted tree. Usually reconstruct unrooted trees. • Mutation mechanisms: – Substitutions – Transpositions – Insertions, Deletions, etc. Will only consider substitutions and assume sequences are aligned. acctaa Put 12/23/2021 acctga Noah Japheth Ham acctga Shem acctga Cush Kannan Mizraim acctga agctga acctga 4

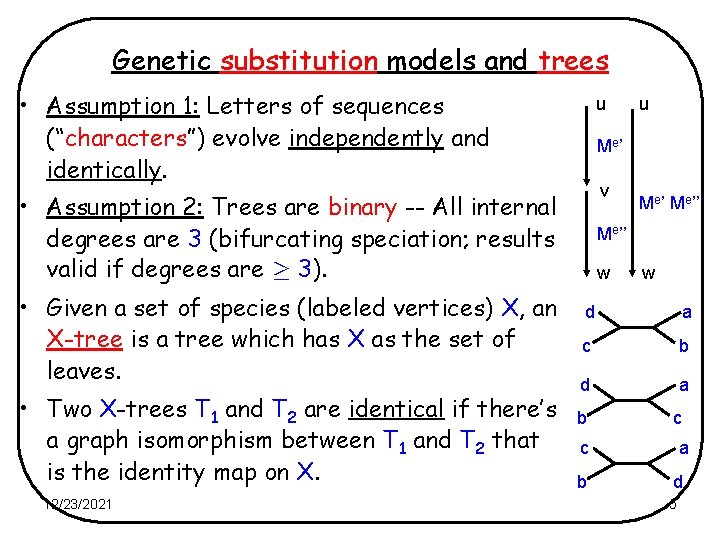
Genetic substitution models and trees • Assumption 1: Letters of sequences (“characters”) evolve independently and identically. u Me ’ v • Assumption 2: Trees are binary -- All internal degrees are 3 (bifurcating speciation; results valid if degrees are ¸ 3). • Given a set of species (labeled vertices) X, an X-tree is a tree which has X as the set of leaves. • Two X-trees T 1 and T 2 are identical if there’s a graph isomorphism between T 1 and T 2 that is the identity map on X. 12/23/2021 u Me’’ w w d a c b d a b c c a b d 5

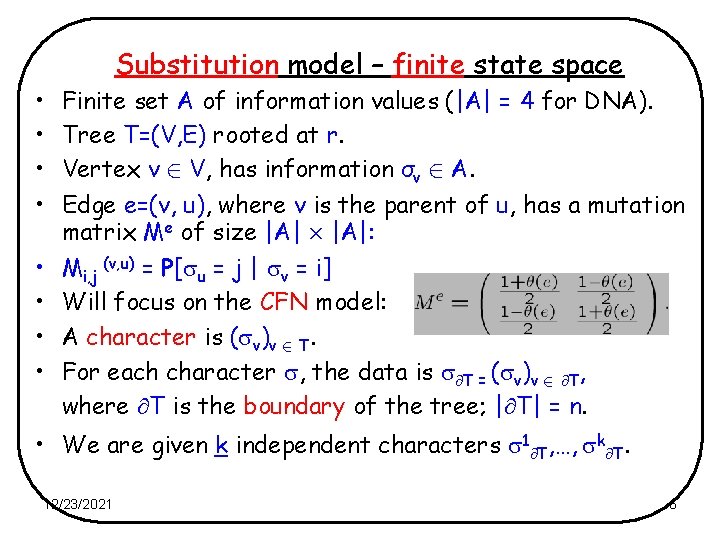
• • Substitution model – finite state space Finite set A of information values (|A| = 4 for DNA). Tree T=(V, E) rooted at r. Vertex v 2 V, has information σv 2 A. Edge e=(v, u), where v is the parent of u, has a mutation matrix Me of size |A| £ |A|: Mi, j (v, u) = P[ u = j | v = i] Will focus on the CFN model: A character is ( v)v 2 T. For each character , the data is T = ( v)v 2 T, where T is the boundary of the tree; | T| = n. • We are given k independent characters 1 T, …, k T. 12/23/2021 6

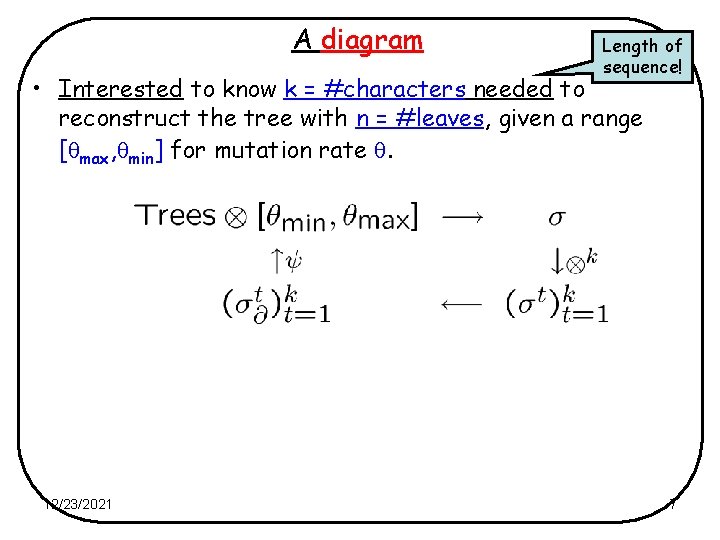
A diagram Length of sequence! • Interested to know k = #characters needed to reconstruct the tree with n = #leaves, given a range [ max, min] for mutation rate . 12/23/2021 7

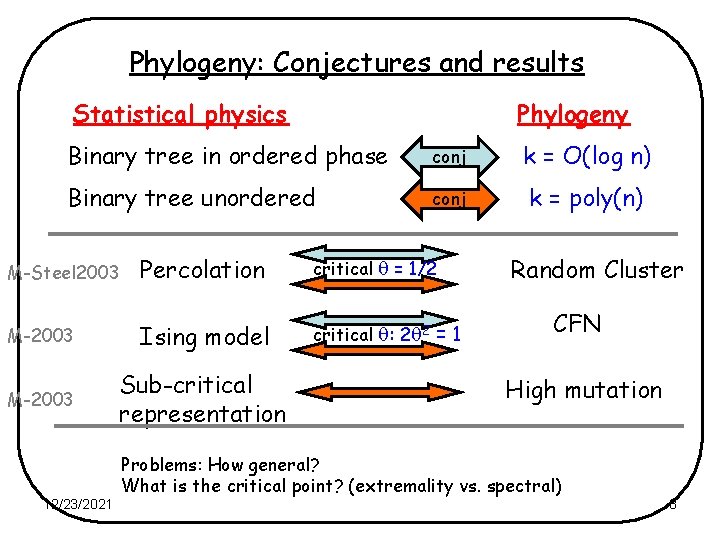
Phylogeny: Conjectures and results Statistical physics Phylogeny Binary tree in ordered phase conj k = O(log n) Binary tree unordered conj k = poly(n) M-Steel 2003 Percolation critical = 1/2 M-2003 Ising model critical : 2 2 = 1 M-2003 Sub-critical representation 12/23/2021 Random Cluster CFN High mutation Problems: How general? What is the critical point? (extremality vs. spectral) 8

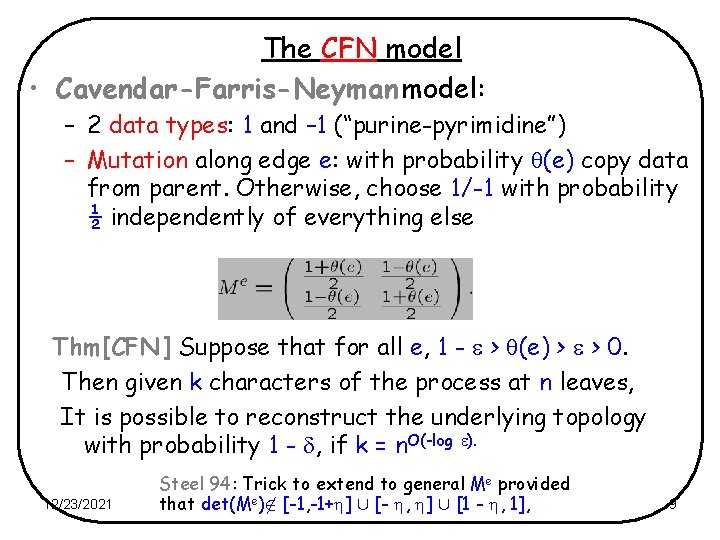
The CFN model • Cavendar-Farris-Neyman model: – 2 data types: 1 and – 1 (“purine-pyrimidine”) – Mutation along edge e: with probability (e) copy data from parent. Otherwise, choose 1/-1 with probability ½ independently of everything else Thm[CFN] Suppose that for all e, 1 - > (e) > > 0. Then given k characters of the process at n leaves, It is possible to reconstruct the underlying topology with probability 1 - , if k = n. O(-log ). 12/23/2021 Steel 94: Trick to extend to general Me provided that det(Me) [-1, -1+ ] [- , ] [1 - , 1], 9
![Phase transition for the CFN model Th 1M 2003 Suppose that n3 Phase transition for the CFN model • Th 1[M 2003]: Suppose that n=3 £](https://slidetodoc.com/presentation_image_h2/2996d4de6919079becc87df885166260/image-10.jpg)
Phase transition for the CFN model • Th 1[M 2003]: Suppose that n=3 £ 2 q and - T is a uniformly chosen (q+1)-level 3 -regular X-tree. – For all e, (e) < , and 2 2 < 1. – Then in order to reconstruct the topology with probability > 0. 1, at least k = (n(-2 log 2( ) - 1)) characters are needed. • Proof: Information theoretic variant of the proof for random cluster model. • Same proof applies to any model for which the reconstruction problem is unsolvable. – more formally, for models for which I( , n) decays exp. fast in n. 12/23/2021 10
![CFN Logarithmic reconstruction Th 2M 2003 If T is an Xtree on n leaves CFN Logarithmic reconstruction Th 2[M 2003]: If T is an X-tree on n leaves](https://slidetodoc.com/presentation_image_h2/2996d4de6919079becc87df885166260/image-11.jpg)
CFN Logarithmic reconstruction Th 2[M 2003]: If T is an X-tree on n leaves s. t. – For all e, min < (e)< max and 2 2 min > 1, max < 1. – Then k = O(log n – log ) characters suffice to reconstruct the topology with probability 1 - . – Need either a “balanced tree” – all leaves at the same distance from a root. – Or, “molecular clock” – (e) = e-t(e), where t(e) is the time interval between the two endpoints of the interval + all leaves are at the same time. 12/23/2021 11
![Main Lemma M 2003 Lemma Suppose that 2 min 2 1 then Main Lemma [M 2003] • Lemma: Suppose that 2 min 2 > 1, then](https://slidetodoc.com/presentation_image_h2/2996d4de6919079becc87df885166260/image-12.jpg)
Main Lemma [M 2003] • Lemma: Suppose that 2 min 2 > 1, then there exists an L, and > 0 such that the CFN model on the binary tree of L levels with – (e) min, for all e not adjacent to ∂T. – (e) min , for all e adjacent to ∂T. satisfies E[σr Maj(σ∂)] . • Roughly, given boundary data of “quality ”, we can reconstruct the root data with “quality ”. • In phylogeny – can treat known pieces of the tree as vertices. • Main problem: how to reconstruct pieces of the tree? 12/23/2021 12

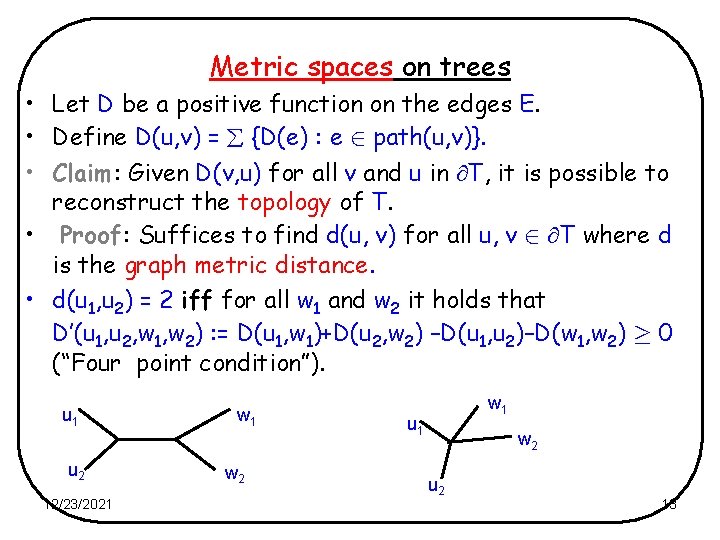
Metric spaces on trees • Let D be a positive function on the edges E. • Define D(u, v) = {D(e) : e 2 path(u, v)}. • Claim: Given D(v, u) for all v and u in T, it is possible to reconstruct the topology of T. • Proof: Suffices to find d(u, v) for all u, v 2 T where d is the graph metric distance. • d(u 1, u 2) = 2 iff for all w 1 and w 2 it holds that D’(u 1, u 2, w 1, w 2) : = D(u 1, w 1)+D(u 2, w 2) –D(u 1, u 2)–D(w 1, w 2) ¸ 0 (“Four point condition”). u 1 u 2 12/23/2021 w 2 w 1 u 1 w 2 u 2 13

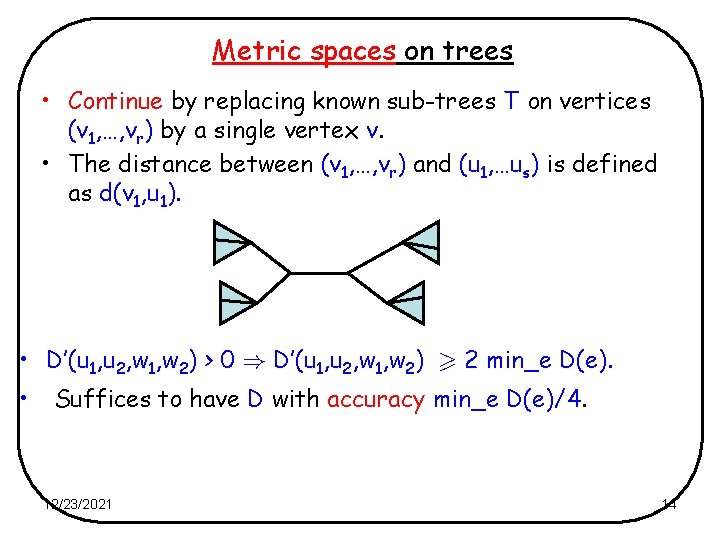
Metric spaces on trees • Continue by replacing known sub-trees T on vertices (v 1, …, vr) by a single vertex v. • The distance between (v 1, …, vr) and (u 1, …us) is defined as d(v 1, u 1). • D’(u 1, u 2, w 1, w 2) > 0 ) D’(u 1, u 2, w 1, w 2) 2 min_e D(e). • Suffices to have D with accuracy min_e D(e)/4. 12/23/2021 14

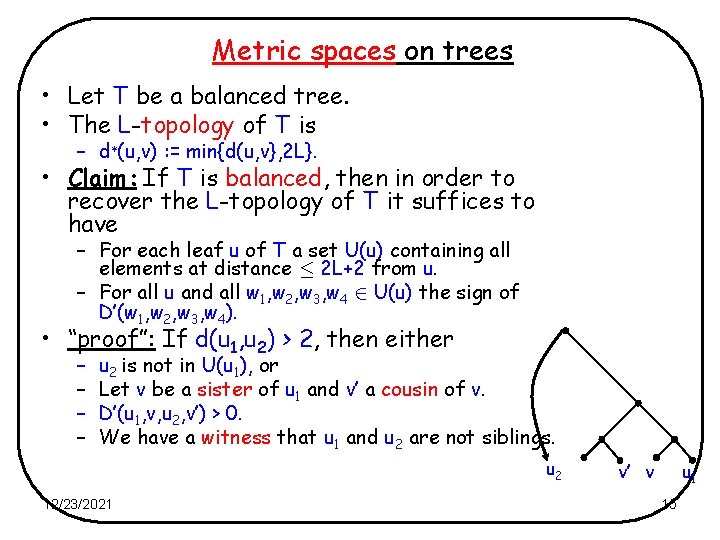
Metric spaces on trees • Let T be a balanced tree. • The L-topology of T is – d¤(u, v) : = min{d(u, v}, 2 L}. • Claim: If T is balanced, then in order to recover the L-topology of T it suffices to have – For each leaf u of T a set U(u) containing all elements at distance · 2 L+2 from u. – For all u and all w 1, w 2, w 3, w 4 2 U(u) the sign of D’(w 1, w 2, w 3, w 4). • “proof”: If d(u 1, u 2) > 2, then either – – u 2 is not in U(u 1), or Let v be a sister of u 1 and v’ a cousin of v. D’(u 1, v, u 2, v’) > 0. We have a witness that u 1 and u 2 are not siblings. u 2 12/23/2021 v’ v u 1 15

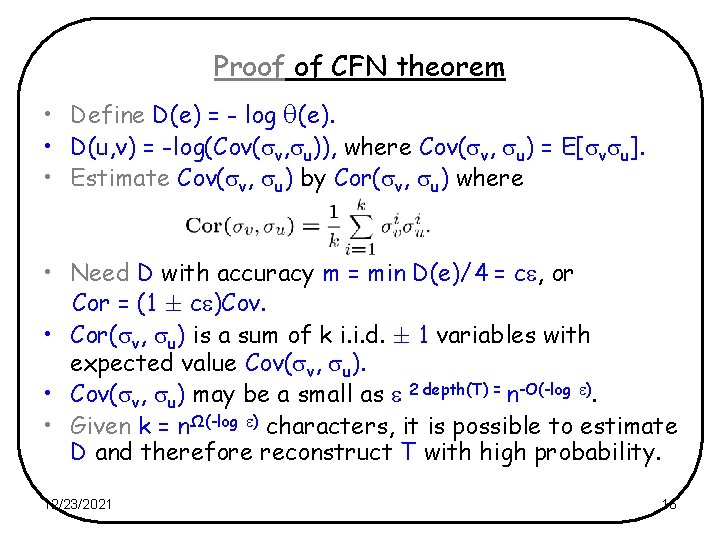
Proof of CFN theorem • Define D(e) = - log (e). • D(u, v) = -log(Cov( v, u)), where Cov( v, u) = E[ v u]. • Estimate Cov( v, u) by Cor( v, u) where • Need D with accuracy m = min D(e)/4 = c , or Cor = (1 c )Cov. • Cor( v, u) is a sum of k i. i. d. 1 variables with expected value Cov( v, u). • Cov( v, u) may be a small as 2 depth(T) = n-O(-log ). • Given k = nΩ(-log ) characters, it is possible to estimate D and therefore reconstruct T with high probability. 12/23/2021 16
![Reconstructing the topology M 2003 The algorithm Repeat the following Reconstruct the Reconstructing the topology [M 2003] • The algorithm: Repeat the following: – Reconstruct the](https://slidetodoc.com/presentation_image_h2/2996d4de6919079becc87df885166260/image-17.jpg)
Reconstructing the topology [M 2003] • The algorithm: Repeat the following: – Reconstruct the topology up to l levels from the boundary using 4 -points method. – For each sample, reconstruct the data l levels from the boundary using majority algorithm. + - - + • Reconstruction near the boundary take O(log n) samples. • By main lemma quality stays above . 12/23/2021 17
![Proving main Lemma Need to estimate Eσr Majσ Estimate has two parts Proving main Lemma • Need to estimate E[σr Maj(σ∂)]. Estimate has two parts: •](https://slidetodoc.com/presentation_image_h2/2996d4de6919079becc87df885166260/image-18.jpg)
Proving main Lemma • Need to estimate E[σr Maj(σ∂)]. Estimate has two parts: • Case 1: For all e adjacent to ∂T, (e) is small. Here we use a perturbation argument, i. e. estimate partial derivatives of E[σr Maj(σ∂)] with respect to various variables (using something like Russo formula). • Case 2: Some e adjacent to ∂T has large (e). Use percolation theory arguments. • Both cases uses isoperimetric estimates for the discrete cube. 12/23/2021 18
 Mossel bay municipality bylaws
Mossel bay municipality bylaws Mossel bay municipality bylaws
Mossel bay municipality bylaws Michel mossel
Michel mossel Dr hurter mossel bay
Dr hurter mossel bay Chapter 26 campbell biology
Chapter 26 campbell biology Chapter 20 phylogeny and the tree of life
Chapter 20 phylogeny and the tree of life Monophyletic grouping
Monophyletic grouping Chapter 26 phylogeny and the tree of life
Chapter 26 phylogeny and the tree of life Chapter 26 phylogeny and the tree of life
Chapter 26 phylogeny and the tree of life Chapter 26 phylogeny and the tree of life
Chapter 26 phylogeny and the tree of life Sister groups on a phylogenetic tree
Sister groups on a phylogenetic tree What is an outgroup in a cladogram
What is an outgroup in a cladogram What is a sister group in phylogeny
What is a sister group in phylogeny Class pterobranchia
Class pterobranchia Barnacle phylogeny
Barnacle phylogeny Biological taxonomy
Biological taxonomy Polytomy
Polytomy Kingdom animalia cladogram
Kingdom animalia cladogram Phylogeny
Phylogeny