Phylogenetic classification of Shiga toxincontaining Escherichia coli Dr






- Slides: 21

Phylogenetic classification of Shiga toxincontaining Escherichia coli Dr. Jim Bono Microbiologist USDA, ARS, US Meat Animal Research Center Meat Safety and Quality Research Unit

Acknowledgments USMARC Dr. Greg Harhay Dr. Mike Clawson Dr. Tim Smith Dr. Jim Keen Sandy Fryda-Bradley Bob Lee Renee Godtel Steve Simcox Linda Flathman Kris Simmerman Randy Bradley Jim Wray Other Collaborators Washington State University Dr. Tom Besser University of Münster Dr. Martina Bielaszewska Dr. Helge Karch Centers for Disease Control and Prevention Dr. Peter Gerner-Smidt Dr. Nancy Strockbine ARS/Western Regional Research Center Dr. Robert Mandrell ARS/Eastern Regional Research Center Dr. Pina Fratamico Food and Drug Administration Dr. Shaohua Zhao Dr. Errol Strain Dr. Marc Allard Public Health Agency of Canada Dr. Roger Johnson Food and Environmental Research Agency Robert Stones Battelle National Biodefense Institute Dr. Adam Phillippy Dr. Sergey Koren

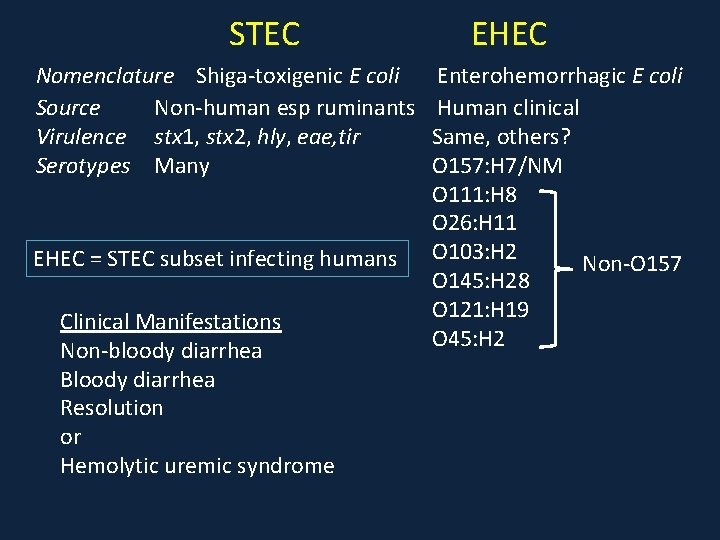
STEC Nomenclature Shiga-toxigenic E coli Source Non-human esp ruminants Virulence stx 1, stx 2, hly, eae, tir Serotypes Many EHEC = STEC subset infecting humans Clinical Manifestations Non-bloody diarrhea Bloody diarrhea Resolution or Hemolytic uremic syndrome EHEC Enterohemorrhagic E coli Human clinical Same, others? O 157: H 7/NM O 111: H 8 O 26: H 11 O 103: H 2 Non-O 157 O 145: H 28 O 121: H 19 O 45: H 2

Shiga toxin-containing Escherichia coli (STEC) • • Zoonotic foodborne human intestinal pathogen Normal, transient, non-pathogenic bovine intestinal microflora Cattle implicated as direct & indirect human infection source Bovine feces assumed to be primary human and bovine contamination & infection source 2/3 of STEC Isolates were O 157: H 7 1/3 of STEC isolates were non-O 157 70% of non-O 157 isolates are from the “Top 6”

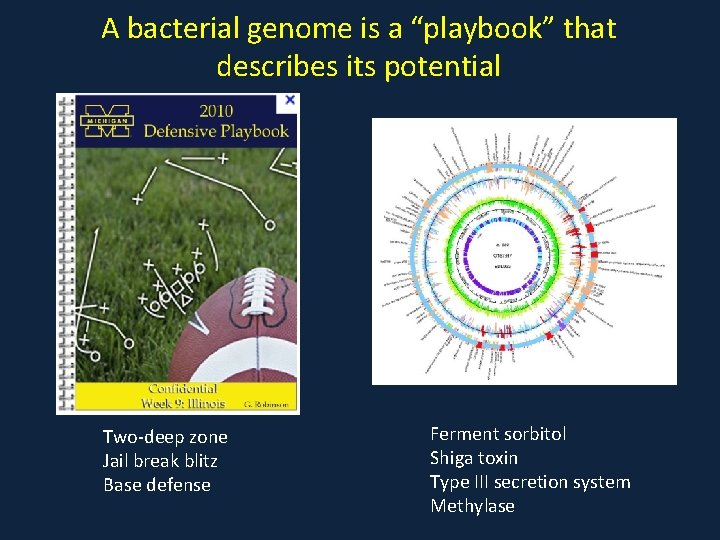
A bacterial genome is a “playbook” that describes its potential Two-deep zone Jail break blitz Base defense Ferment sorbitol Shiga toxin Type III secretion system Methylase

Family Tree

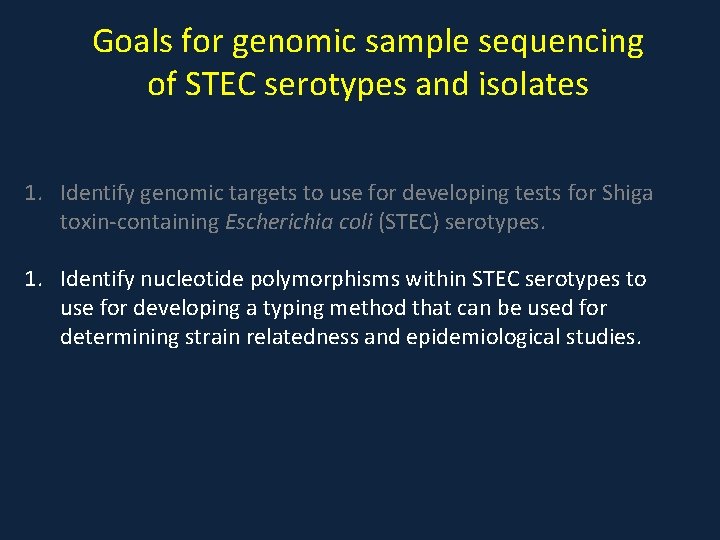
Goals for genomic sample sequencing of STEC serotypes and isolates 1. Identify genomic targets to use for developing tests for Shiga toxin-containing Escherichia coli (STEC) serotypes. 1. Identify nucleotide polymorphisms within STEC serotypes to use for developing a typing method that can be used for determining strain relatedness and epidemiological studies.

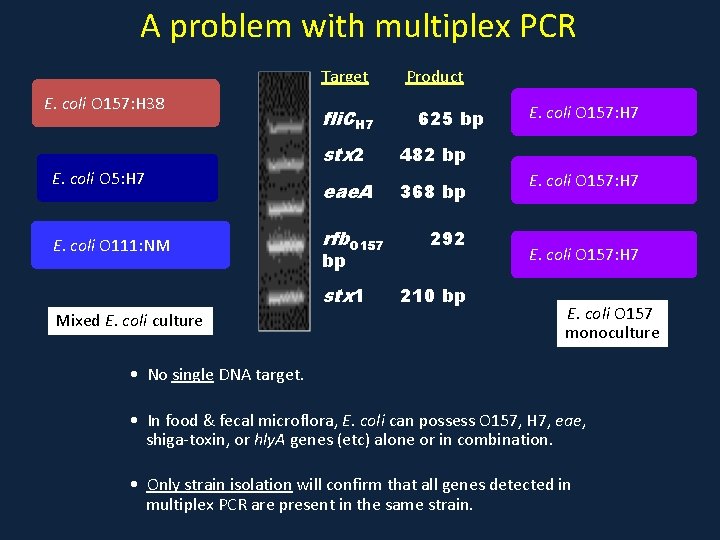
A problem with multiplex PCR Target E. coli O 157: H 38 E. coli O 5: H 7 E. coli O 111: NM fli. CH 7 625 bp stx 2 482 bp eae. A 368 bp rfb. O 157 bp stx 1 Mixed E. coli culture Product 292 210 bp E. coli O 157: H 7 E. coli O 157 monoculture • No single DNA target. • In food & fecal microflora, E. coli can possess O 157, H 7, eae, shiga-toxin, or hly. A genes (etc) alone or in combination. • Only strain isolation will confirm that all genes detected in multiplex PCR are present in the same strain.

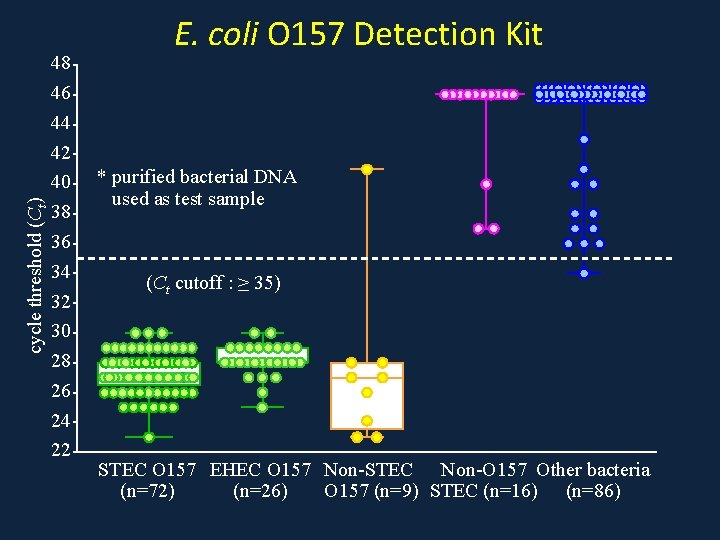
cycle threshold (Ct) 48 46 44 42 40 38 36 34 32 30 28 26 24 22 E. coli O 157 Detection Kit * purified bacterial DNA used as test sample (Ct cutoff : ≥ 35) STEC O 157 EHEC O 157 Non-STEC Non-O 157 Other bacteria (n=72) (n=26) O 157 (n=9) STEC (n=16) (n=86)

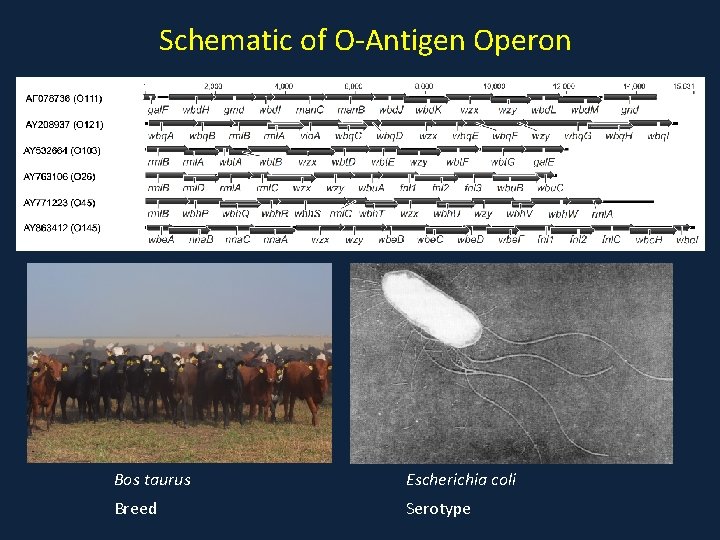
Schematic of O-Antigen Operon Bos taurus Escherichia coli Breed Serotype

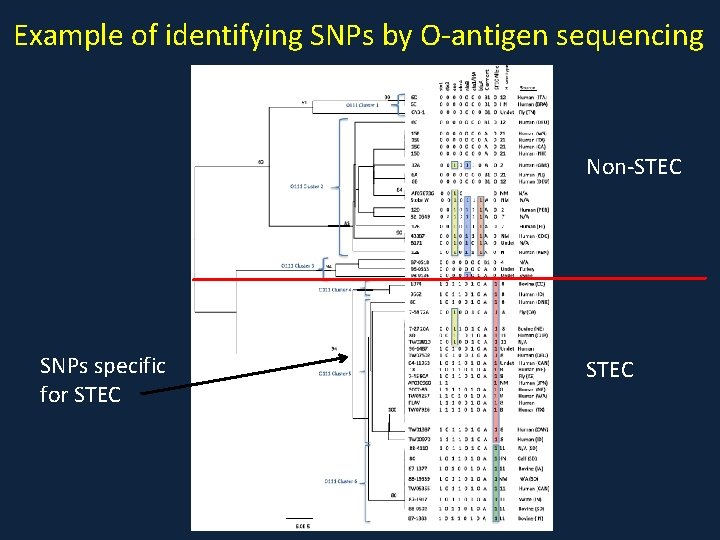
Example of identifying SNPs by O-antigen sequencing Non-STEC SNPs specific for STEC

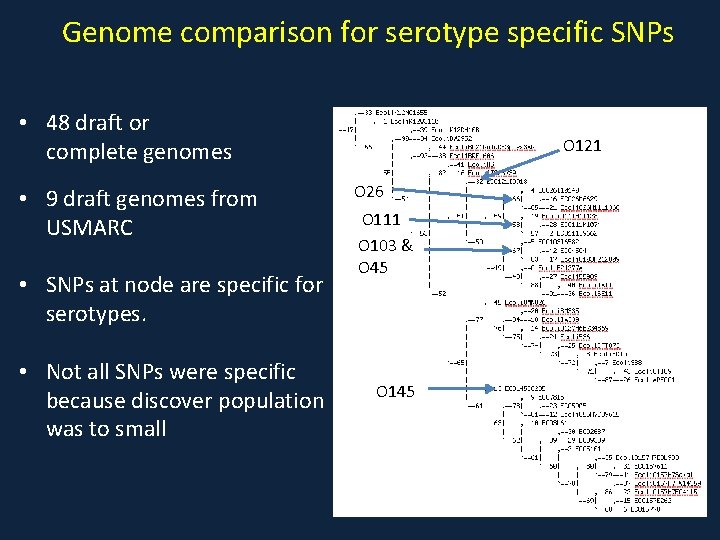
Genome comparison for serotype specific SNPs • 48 draft or complete genomes • 9 draft genomes from USMARC • SNPs at node are specific for serotypes. • Not all SNPs were specific because discover population was to small O 121 O 26 O 111 O 103 & O 45 O 145

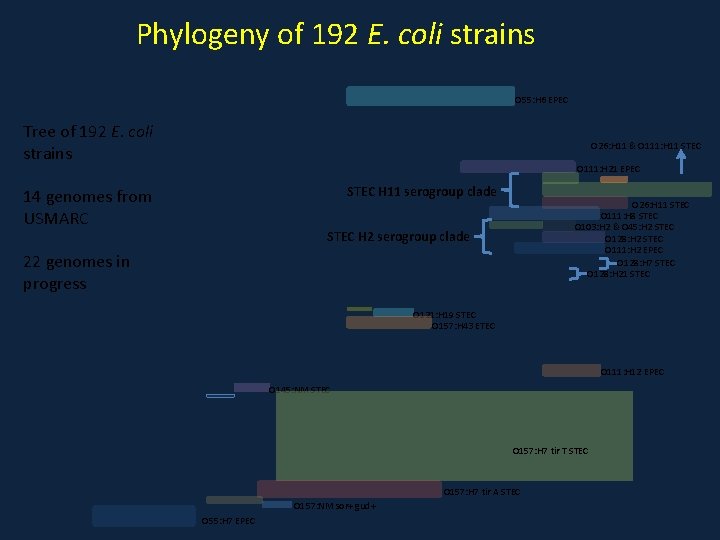
Phylogeny of 192 E. coli strains O 55: H 6 EPEC Tree of 192 E. coli strains O 26: H 11 & O 111: H 11 STEC O 111: H 21 EPEC STEC H 11 serogroup clade 14 genomes from USMARC O 26: H 11 STEC O 111: H 8 STEC O 103: H 2 & O 45: H 2 STEC O 128: H 2 STEC O 111: H 2 EPEC O 128: H 7 STEC O 128: H 21 STEC H 2 serogroup clade 22 genomes in progress O 121: H 19 STEC O 157: H 43 ETEC O 111: H 12 EPEC O 145: NM STEC O 157: H 7 tir T STEC O 157: H 7 tir A STEC O 157: NM sor+ gud+ O 55: H 7 EPEC

Accomplishments O-antigen operons have SNPs that can be used to differentiate STEC from non-STEC strains. Serotype specific SNPs can be identified through genome comparison. Impact Serotype specific SNPs from the O-antigen sequencing project have been licensed and are being used in a STEC detection and identification system. This system was recently award a letter of no objection by FSIS, which allows companies to use this system to comply with recently implemented regulations regarding testing for 6 STEC non-O 157 serogroups, in addition to STEC O 157: H 7.

Goals for genomic sample sequencing of STEC serotypes and isolates 1. Identify genomic targets to use for developing tests for Shiga toxin-containing Escherichia coli (STEC) serotypes. 1. Identify nucleotide polymorphisms within STEC serotypes to use for developing a typing method that can be used for determining strain relatedness and epidemiological studies.

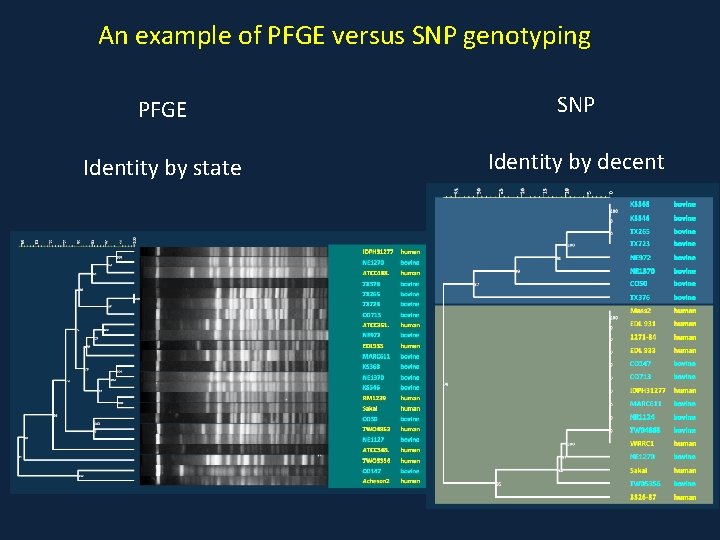
An example of PFGE versus SNP genotyping PFGE SNP Identity by state Identity by decent

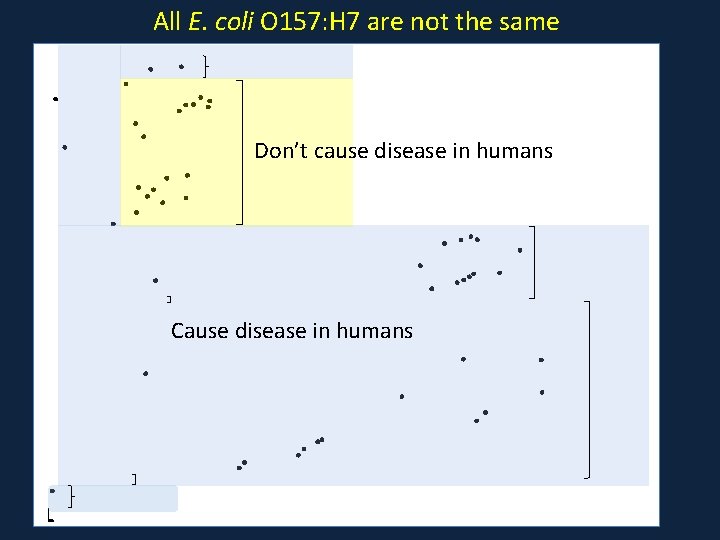
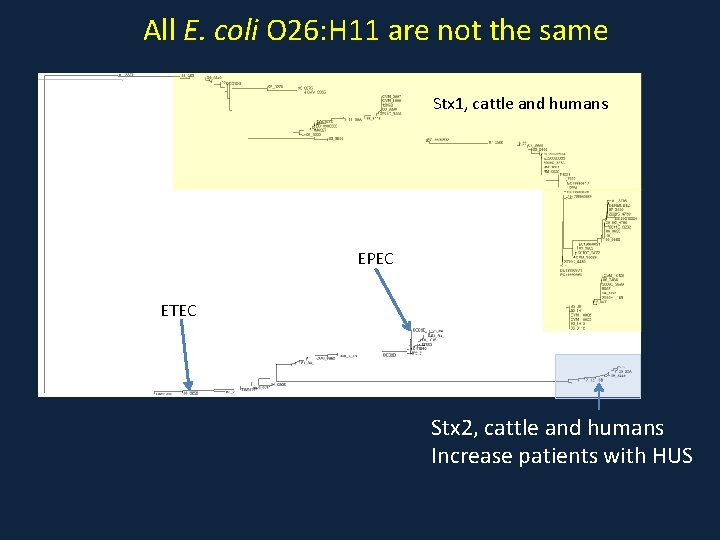
All E. coli O 157: H 7 are not the same Don’t cause disease in humans Cause disease in humans

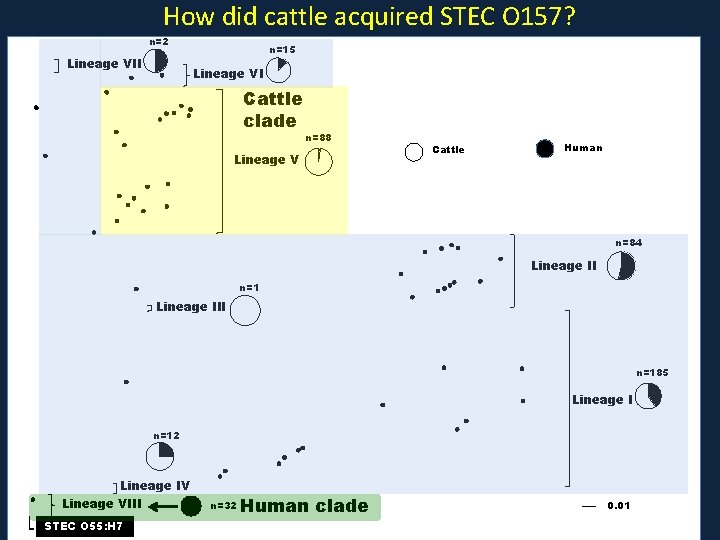
How did cattle acquired STEC O 157? n=2 Lineage VII n=15 Lineage VI Cattle clade n=88 Lineage V Cattle Human n=84 Lineage II n=1 Lineage III n=185 Lineage I n=12 Lineage IV Lineage VIII STEC O 55: H 7 n=32 Human clade 0. 01

All E. coli O 26: H 11 are not the same Stx 1, cattle and humans EPEC ETEC Stx 2, cattle and humans Increase patients with HUS

Accomplishments A set of nucleotide polymorphisms has been developed for detecting STEC O 157 and O 26 genetic subtypes through identity-by descent. STEC O 157 evolution has been redefined with this set of polymorphisms. This is the first large scale SNP discovery and analysis of relatedness for serogroup O 26 Impact CDC is using STEC O 157 SNPs in forming a group of SNPs to genotype EHEC O 157 strains.

Questions?
 Escherichia coli unicellular or multicellular
Escherichia coli unicellular or multicellular Genetic toggle switch
Genetic toggle switch Escherichia coli
Escherichia coli Escherichia coli reino al que pertenece
Escherichia coli reino al que pertenece Escherichia coli mobile ou immobile
Escherichia coli mobile ou immobile Tsi agar
Tsi agar Coprocultivo diagrama de flujo
Coprocultivo diagrama de flujo Bacteriophage virus
Bacteriophage virus Genul escherichia
Genul escherichia Escherichia spp
Escherichia spp The importance of microbes
The importance of microbes Branching key example
Branching key example Lake of the ozarks e coli
Lake of the ozarks e coli Epiploicum
Epiploicum E coli tca cycle
E coli tca cycle Rizomelie
Rizomelie Balantidium coli
Balantidium coli Define enteric bacteria
Define enteric bacteria Bulbus duodeni
Bulbus duodeni Fornix gastricus
Fornix gastricus Stefni coli
Stefni coli Api test microbiology
Api test microbiology