Phen Code Linking Human Mutations to Phenotype Phen




- Slides: 16

Phen. Code Linking Human Mutations to Phenotype

Phen. Code • Brings the deep information on genotypes and phenotypes in locus specific databases (LSDBs) into a common database • Uses genome sequence coordinates • Allows data to be displayed in genome browsers and compared analyzed with respect to other genome data. • Collaboration among many LSDBs, Human Genome Variation Society and genome browsers

23 (142) LSDBs Over 17, 000 mutations Swiss-Prot / Uni. Prot Over 23, 000 mutations

Access to data in Phen. Code • Home Page www. bx. psu. edu/phencode – Query page – History page • Output can be viewed as – a custom track at UCSC Genome Browser • (then Table Browser and Genome Graphs) – A custom track at Ensembl – Custom track files for other Genome Browsers – As tab delimited text for downloads

“What LSDB variants are NOT in db. SNP and ARE in conserved regions? ” • Find substitutions in Phen. Code / Locus Variants track • Remove entries that are already in db. SNP • Remove entries that are not intersected by the most conserved track (Phast. Cons) • View results as coverage on genome and details of an single variant

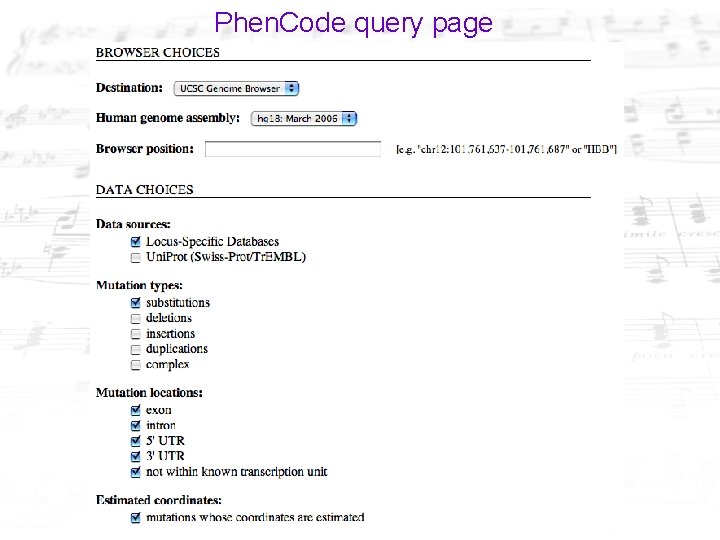
Phen. Code query page

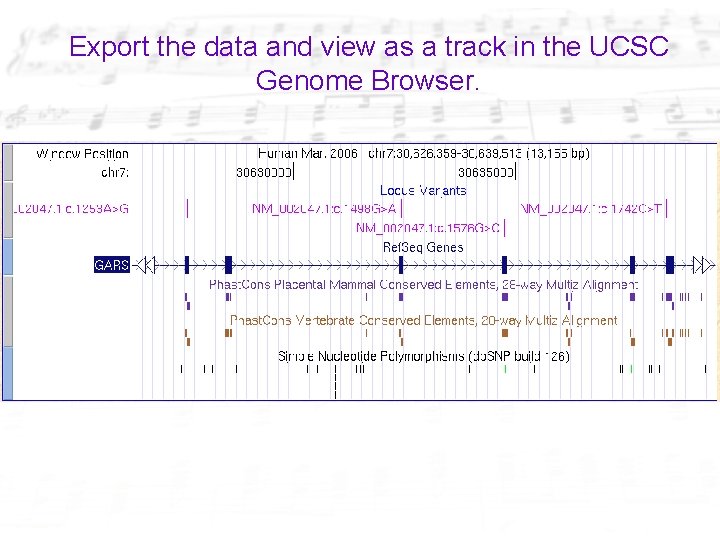
Export the data and view as a track in the UCSC Genome Browser.

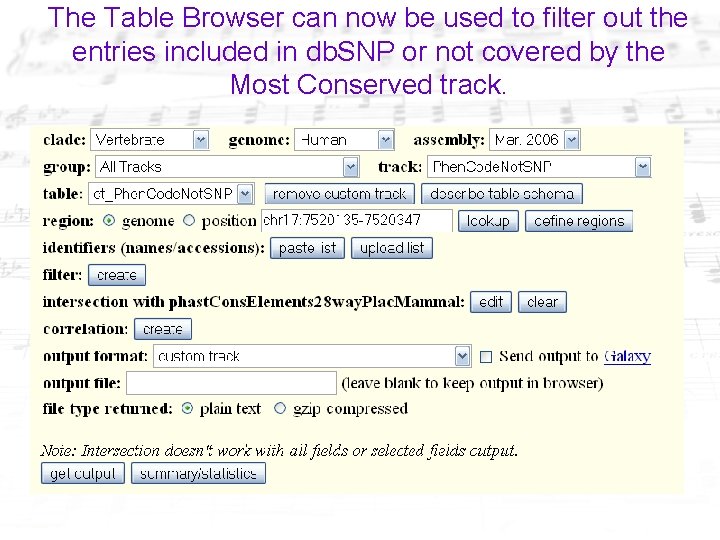
The Table Browser can now be used to filter out the entries included in db. SNP or not covered by the Most Conserved track.

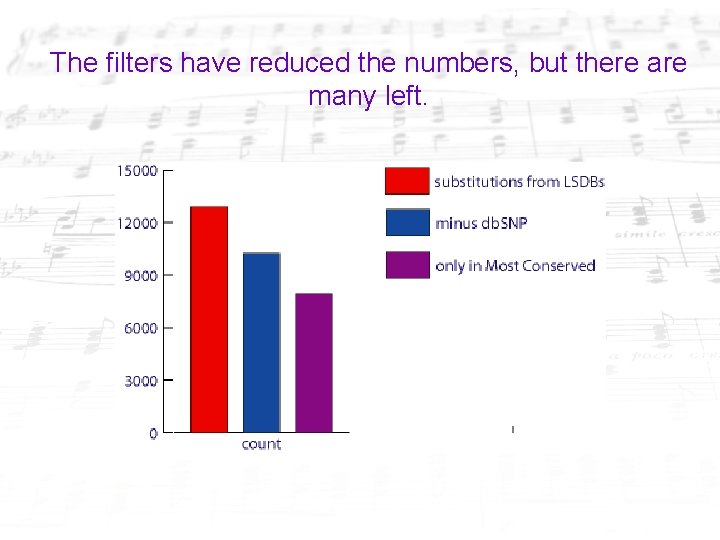
The filters have reduced the numbers, but there are many left.

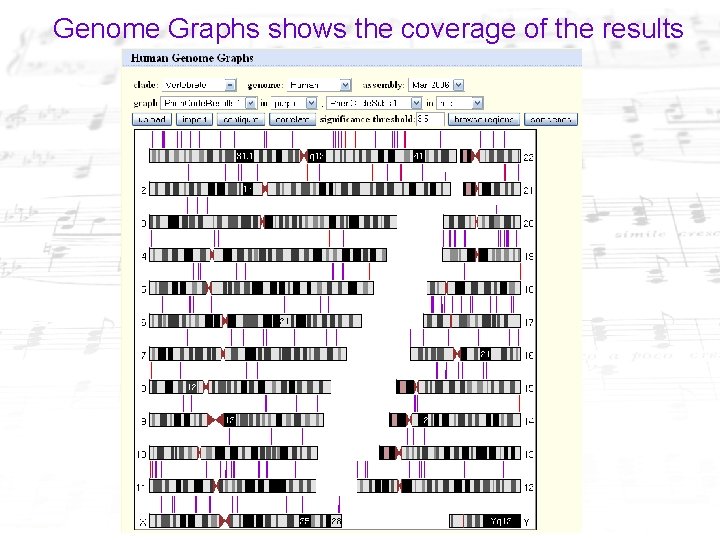
Genome Graphs shows the coverage of the results

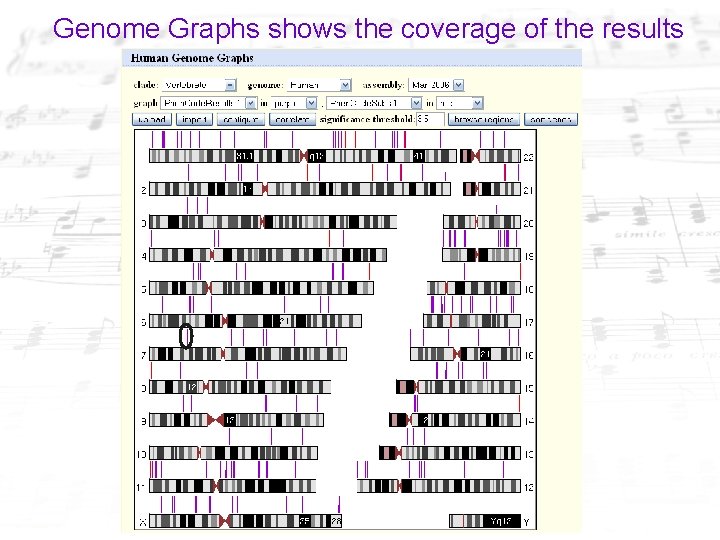
Genome Graphs shows the coverage of the results

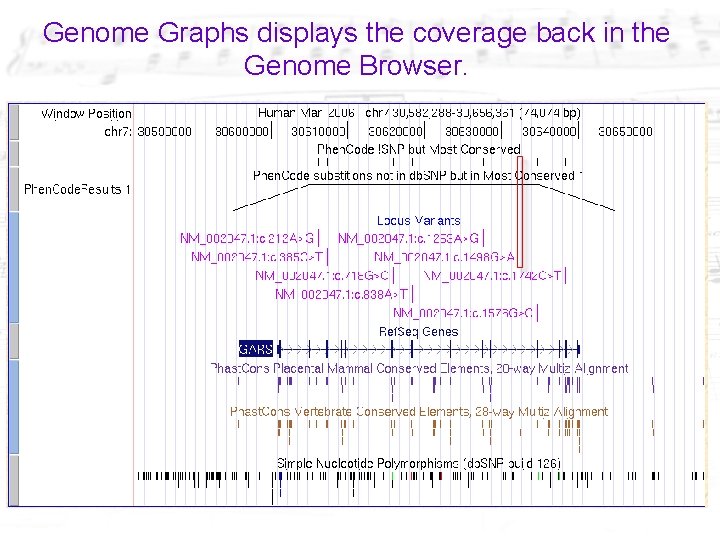
Genome Graphs displays the coverage back in the Genome Browser.

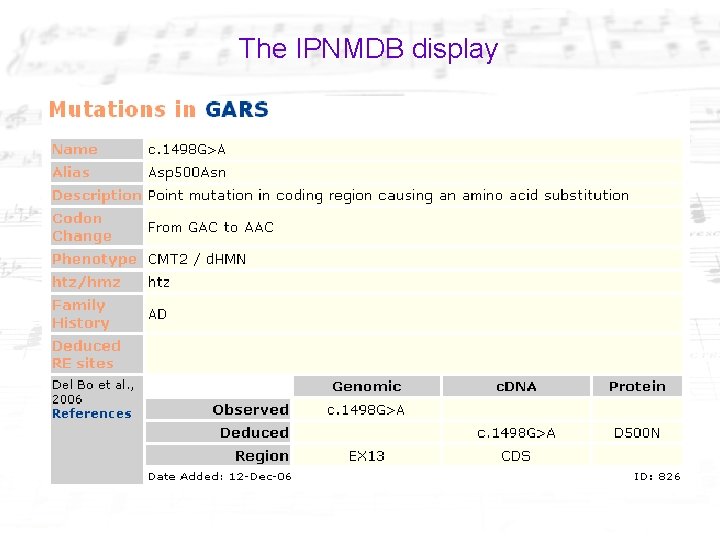
A summary can be found at the Phen. Code site and more details at the source LSDB. CMT 2 = Charcot-Marie-Tooth disease - OMIM 600882, 118210, 605588, 605589, 601472 most common disorder of the peripheral nervous system. (described in 1886) d. HMN = distal Hereditary Motor Neuropathies - OMIM 158590, 600794, …

The IPNMDB display

Conclusion • The deep annotations from LSDBs compliment the broad annotations from db. SNP • The data from Phen. Code is public and available in genome browsers and as text. – www. bx. psu. edu/phencode

Acknowledgements • Work was supported by – NIH grant HG 002238 (Miller) – NIH grant DK 65806 (Hardison) – NHGRI grant 1 P 41 HG 02371 (Kent)