Network Dynamics and Cell Physiology John J Tyson



- Slides: 34

Network Dynamics and Cell Physiology John J. Tyson Dept. Biological Sciences Virginia Tech

Collaborators Budapest Univ. Techn. & Econ. Bela Novak Attila Csikasz-Nagy Andrea Ciliberto Virginia Tech Kathy Chen Dorjsuren Battogtokh Funding James S. Mc. Donnell Foundation DARPA

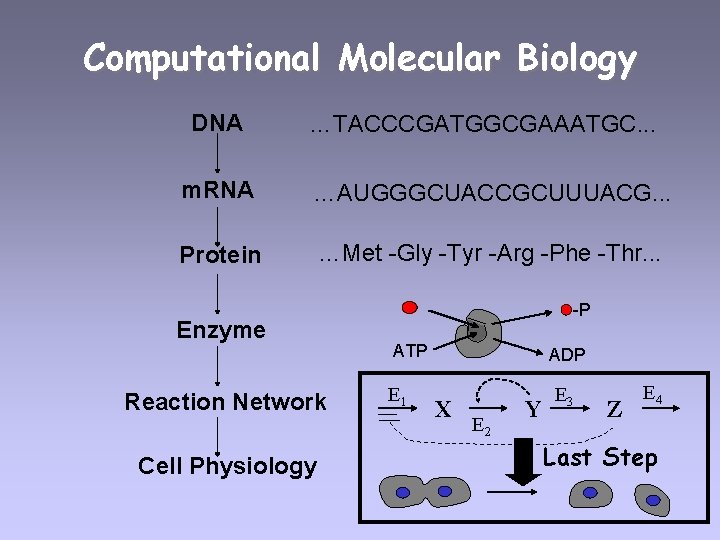
Computational Molecular Biology DNA …TACCCGATGGCGAAATGC. . . m. RNA …AUGGGCUACCGCUUUACG. . . Protein …Met -Gly -Tyr -Arg -Phe -Thr. . . Enzyme Reaction Network Cell Physiology -P ATP E 1 ADP X E 2 Y E 3 Z E 4 Last Step

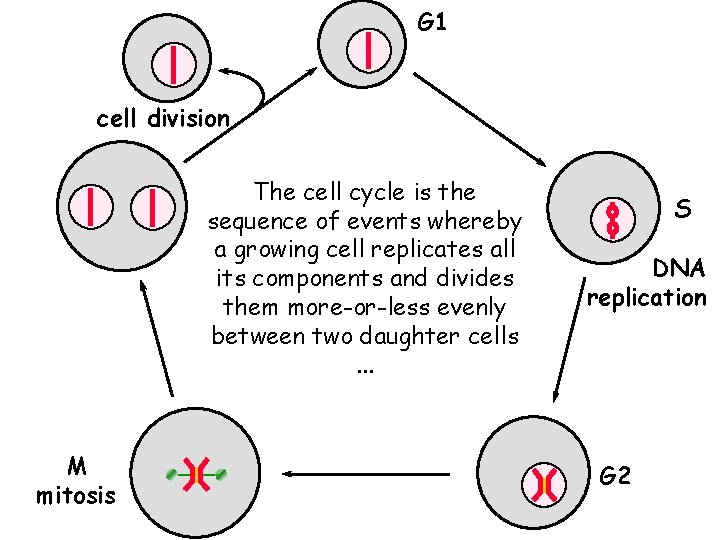
G 1 cell division The cell cycle is the sequence of events whereby a growing cell replicates all its components and divides them more-or-less evenly between two daughter cells. . . M mitosis S DNA replication G 2

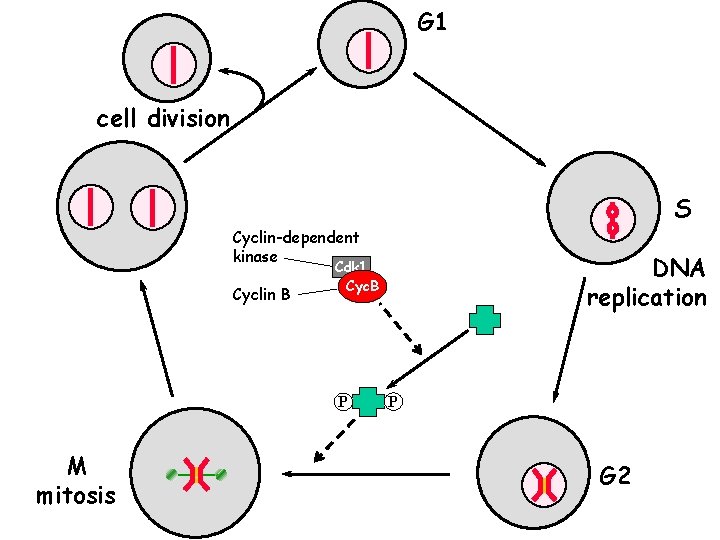
G 1 cell division S Cyclin-dependent kinase Cyclin B P M mitosis DNA replication Cdk 1 Cyc. B P G 2

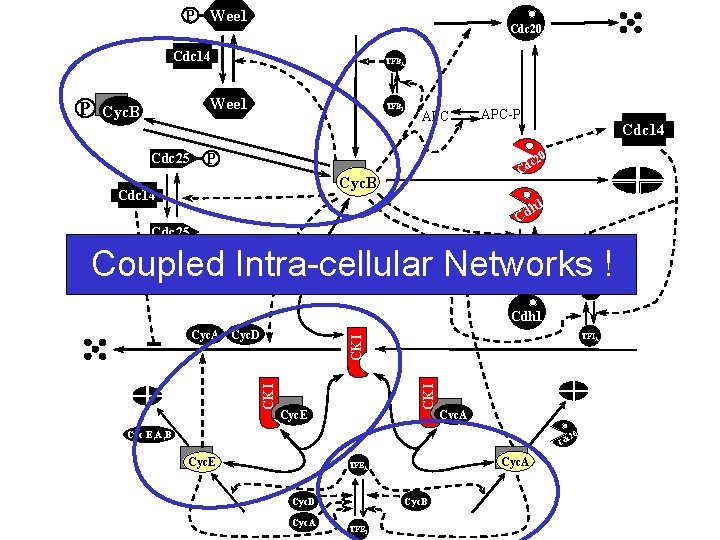
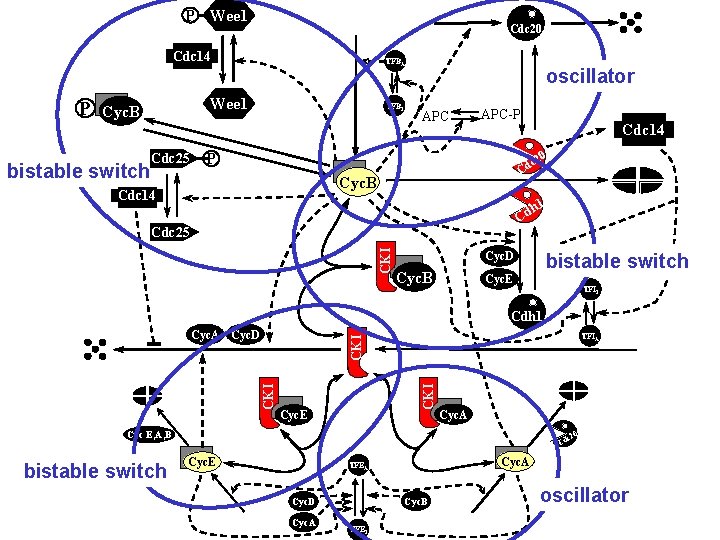
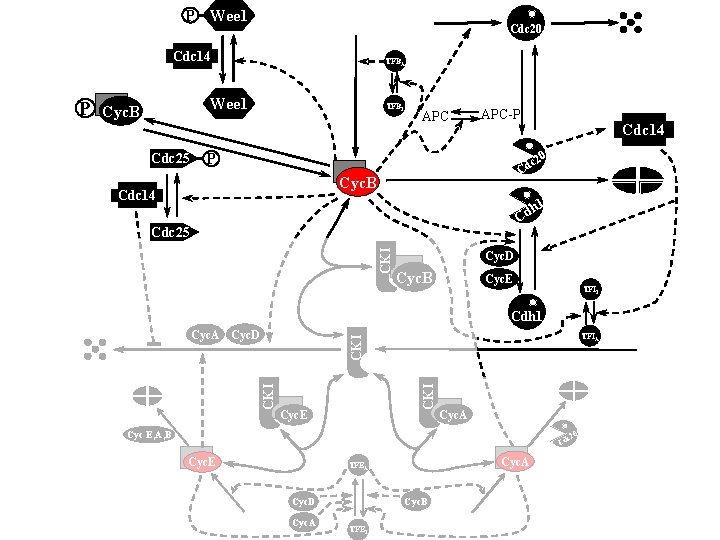
P Wee 1 Cdc 20 Cdc 14 P TFBA Wee 1 Cyc. B Cdc 25 TFBI APC-P Cdc 14 0 P c 2 Cd Cyc. B Cdc 14 h 1 Cd Cdc 25 CKI Coupled Intra-cellular Cyc. B Networks ! Cyc. D Cyc. E TFII Cdh 1 TFIA CKI CKI Cyc. A Cyc. D Cyc. E Cyc. A Cyc E, A, B 0 c 2 Cd Cyc. E Cyc. A TFEA Cyc. B Cyc. D Cyc. A TFEI

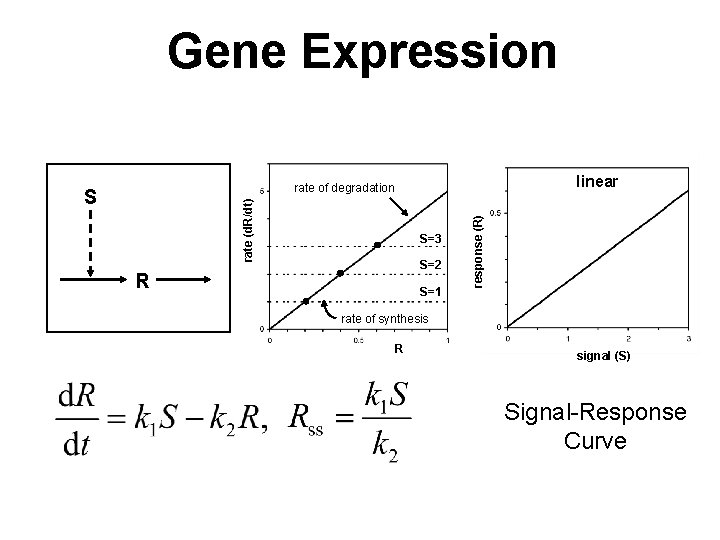
Gene Expression linear rate (d. R/dt) S S=3 S=2 R S=1 response (R) rate of degradation rate of synthesis R signal (S) Signal-Response Curve

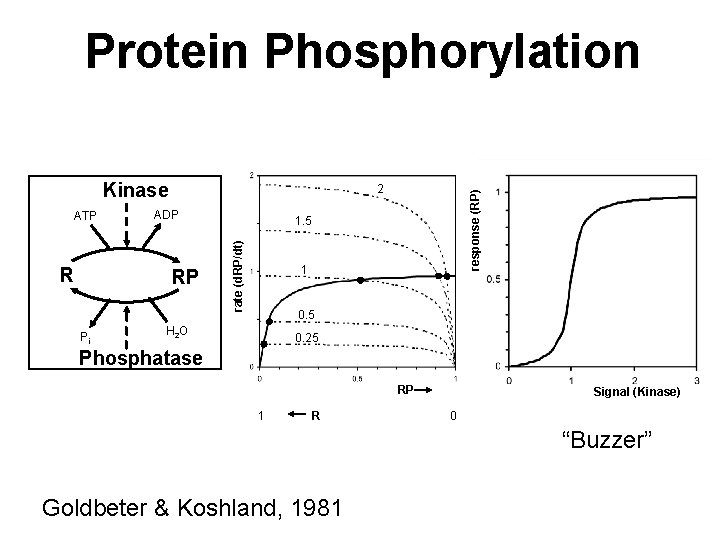
Protein Phosphorylation Kinase RP Pi response (RP) R ADP 1. 5 rate (d. RP/dt) ATP 2 1 0. 5 H 2 O 0. 25 Phosphatase RP 1 R Signal (Kinase) 0 “Buzzer” Goldbeter & Koshland, 1981

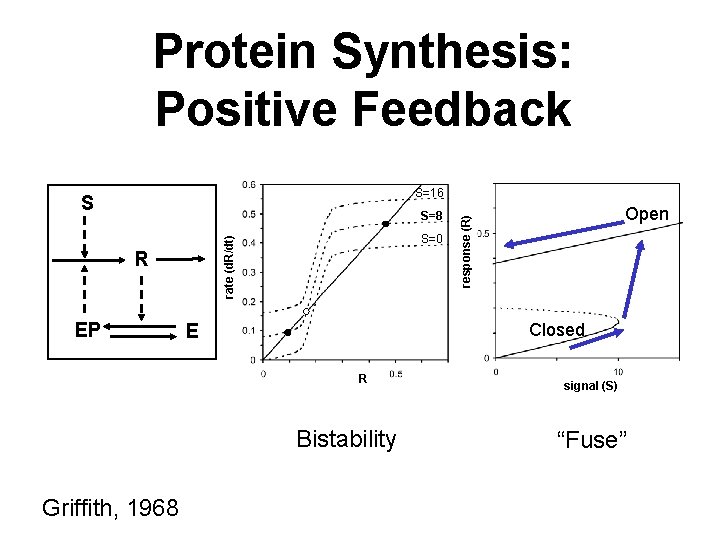
Protein Synthesis: Positive Feedback S=16 S=8 rate (d. R/dt) R EP S=0 Closed E R Bistability Griffith, 1968 Open response (R) S signal (S) “Fuse”

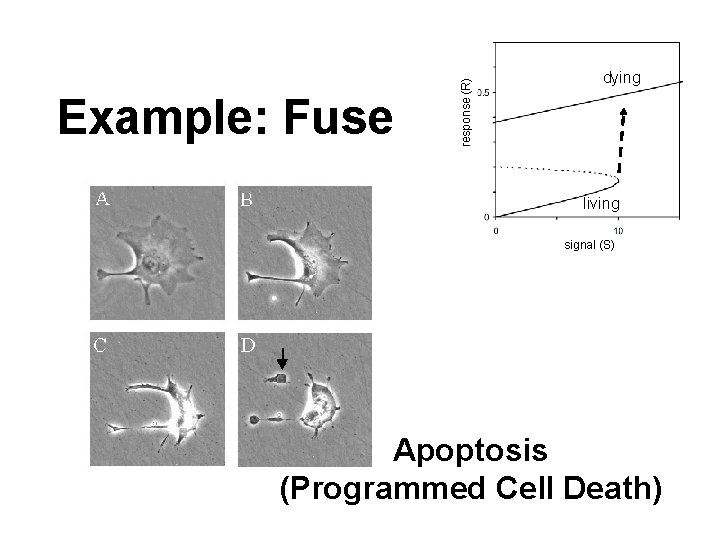
response (R) Example: Fuse dying living signal (S) Apoptosis (Programmed Cell Death)

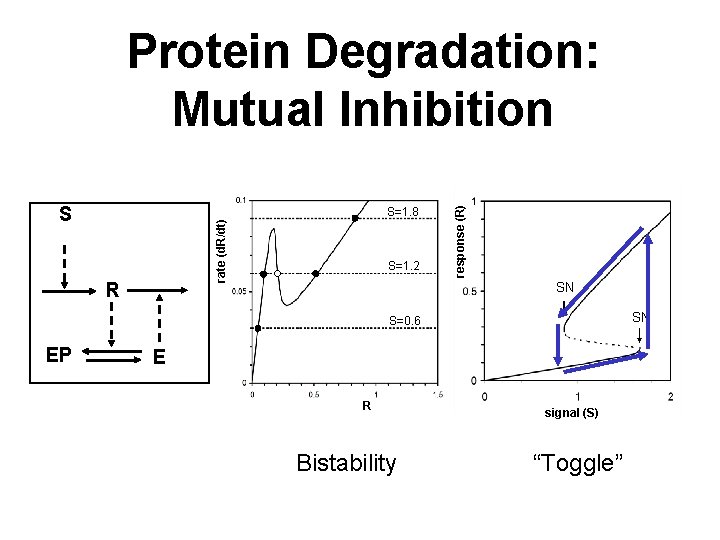
S rate (d. R/dt) S=1. 8 R S=1. 2 response (R) Protein Degradation: Mutual Inhibition SN SN S=0. 6 EP E R Bistability signal (S) “Toggle”

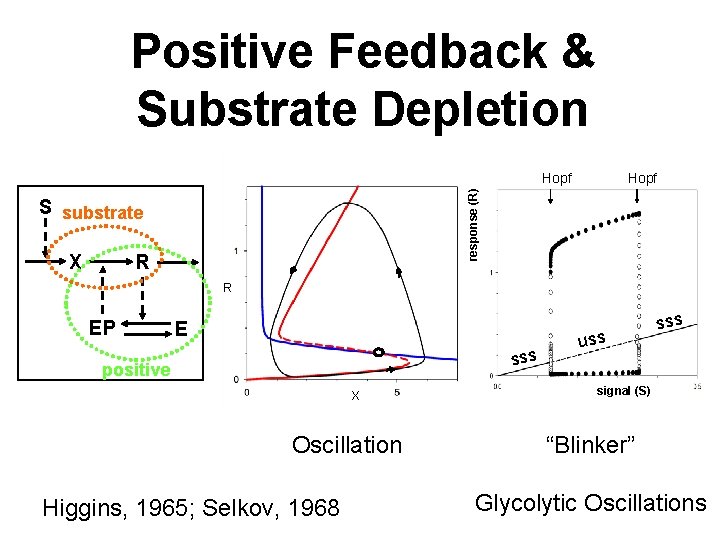
Positive Feedback & Substrate Depletion Hopf response (R) Hopf S substrate X R R EP E sss positive X Oscillation Higgins, 1965; Selkov, 1968 uss signal (S) “Blinker” Glycolytic Oscillations

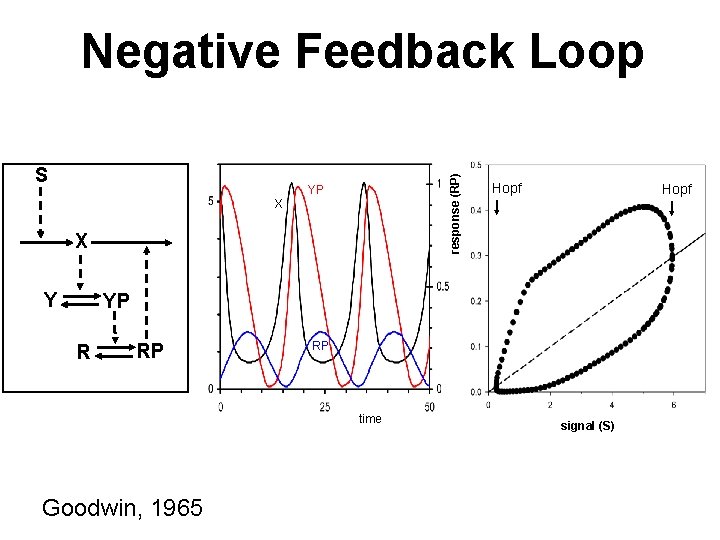
S response (RP) Negative Feedback Loop YP X X Y Hopf YP R RP RP time Goodwin, 1965 signal (S)

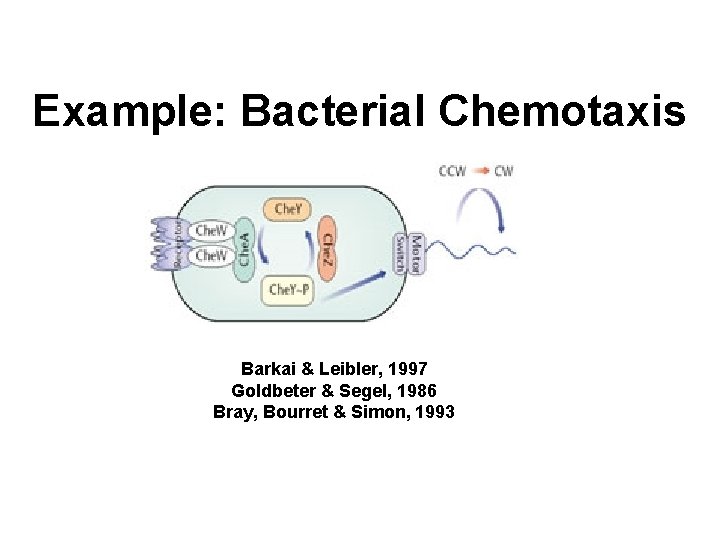
Example: Bacterial Chemotaxis Barkai & Leibler, 1997 Goldbeter & Segel, 1986 Bray, Bourret & Simon, 1993

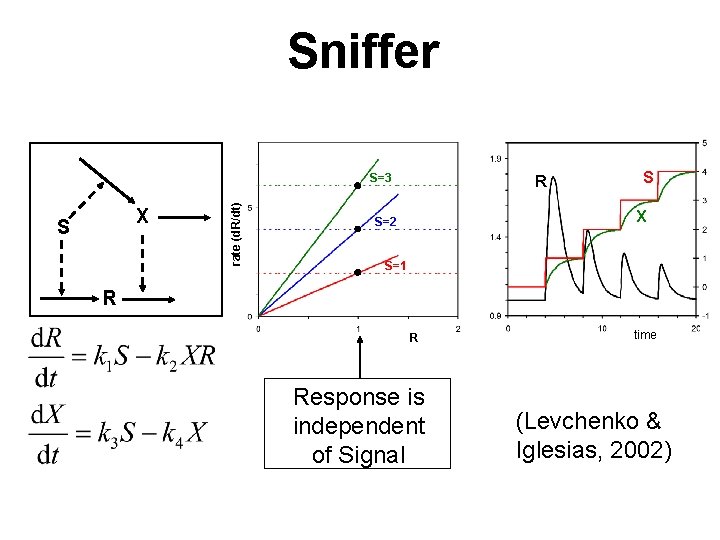
Sniffer X S rate (d. R/dt) S=3 R S X S=2 S=1 R R Response is independent of Signal time (Levchenko & Iglesias, 2002)

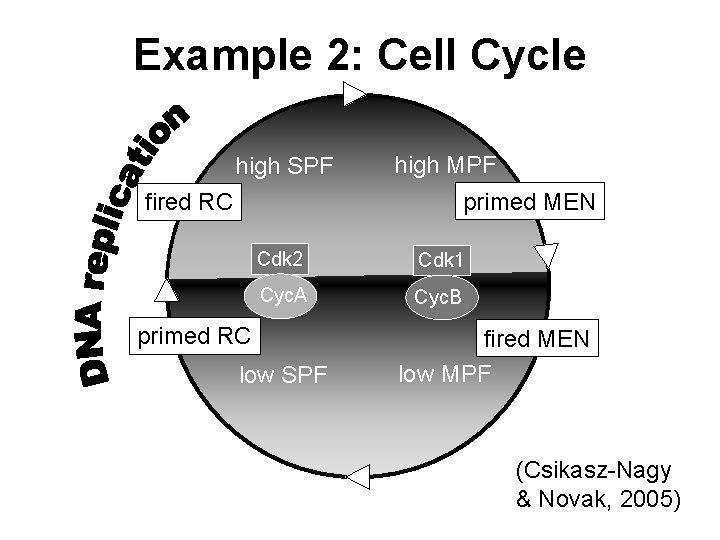
Example 2: Cell Cycle high SPF high MPF primed MEN fired RC Cdk 2 Cdk 1 Cyc. A Cyc. B primed RC low SPF fired MEN low MPF (Csikasz-Nagy & Novak, 2005)

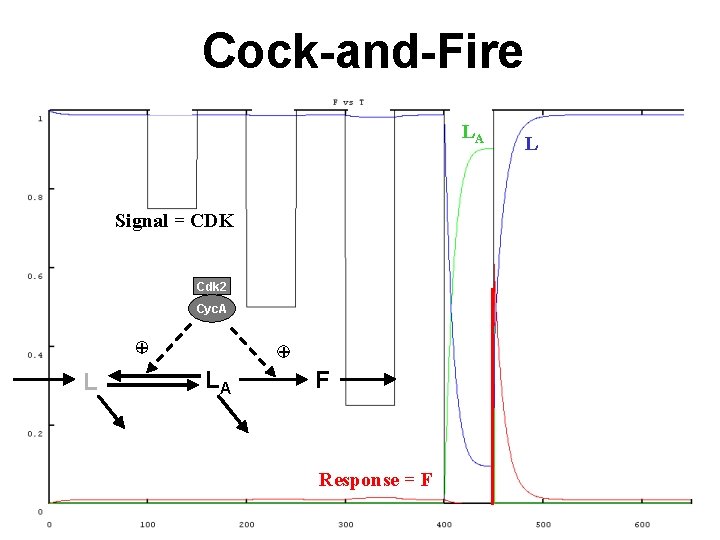
Cock-and-Fire LA Signal = CDK Cdk 2 Cyc. A + LA F Response = F L

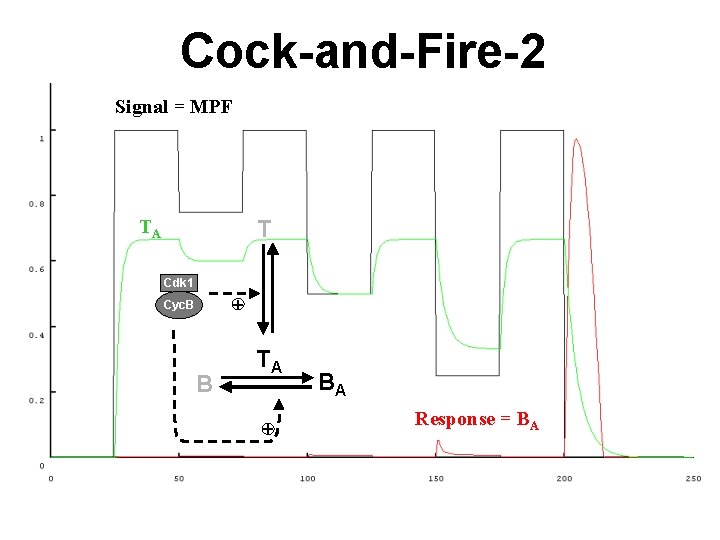
Cock-and-Fire-2 Signal = MPF TA T Cdk 1 + Cyc. B B TA + BA Response = BA

P Wee 1 Cdc 20 Cdc 14 P Wee 1 Cyc. B bistable switch TFBA Cdc 25 TFBI oscillator APC-P Cdc 14 0 P c 2 Cd Cyc. B Cdc 14 h 1 Cd CKI Cdc 25 Cyc. D Cyc. B bistable switch Cyc. E TFII Cdh 1 TFIA CKI CKI Cyc. A Cyc. D Cyc. E Cyc. A Cyc E, A, B bistable switch 0 c 2 Cd Cyc. E Cyc. A TFEA Cyc. B Cyc. D Cyc. A TFEI oscillator

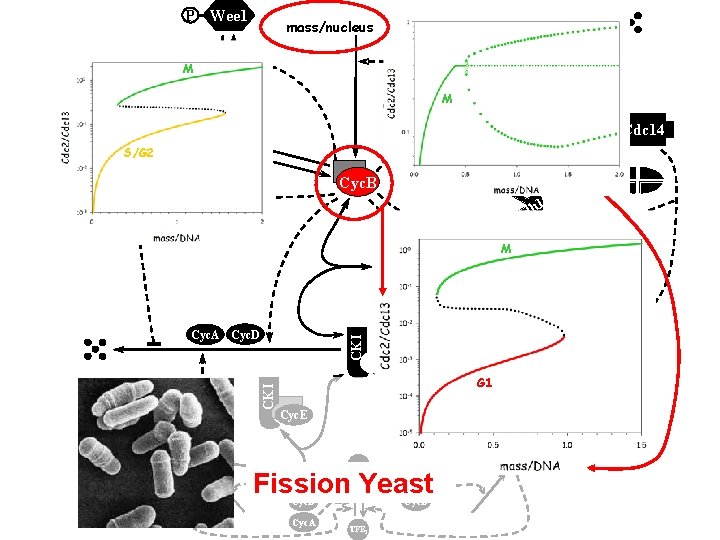
P Wee 1 mass/nucleus Cdc 14 TFBA M Wee 1 Cyc. B S/G 2 Cdc 25 TFBI M APC-P Cdc 14 0 P c 2 Cd Cyc. B Cdc 14 h 1 Cd CKI Cdc 25 M Cyc. D Cyc. B Cyc. E TFII Cdh 1 TFIA CKI Cyc. A Cyc. D CKI P Cdc 20 Cyc. E G 1 Cyc. A Cyc E, A, B 0 c 2 Cd Cyc. E TFEA Fission Yeast Cyc. B Cyc. D Cyc. A TFEI Cyc. A

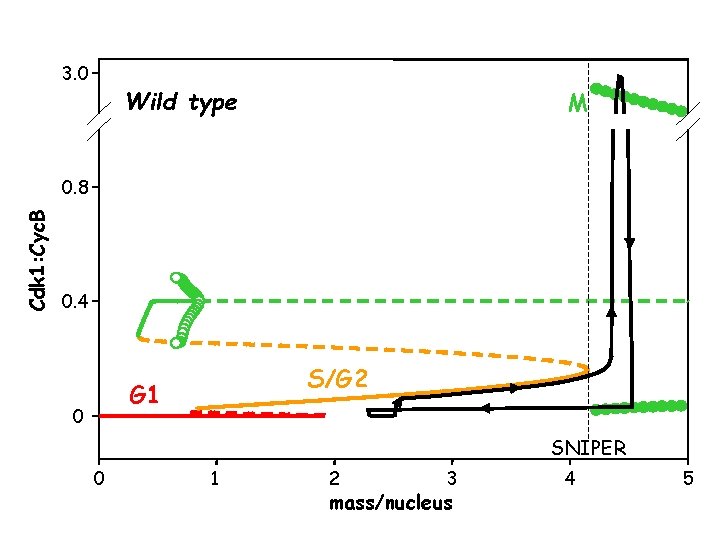
3. 0 Wild type M Cdk 1: Cyc. B 0. 8 0. 4 S/G 2 G 1 0 SNIPER 0 1 2 3 mass/nucleus 4 5

Nature, Vol, 256, No. 5518, pp. 547 -551, August 14, 1975 Genetic control of cell size at cell division in yeast Paul Nurse Department of Zoology, West Mains Road, Edinburgh EH 9 3 JT, UK wild-type wee 1

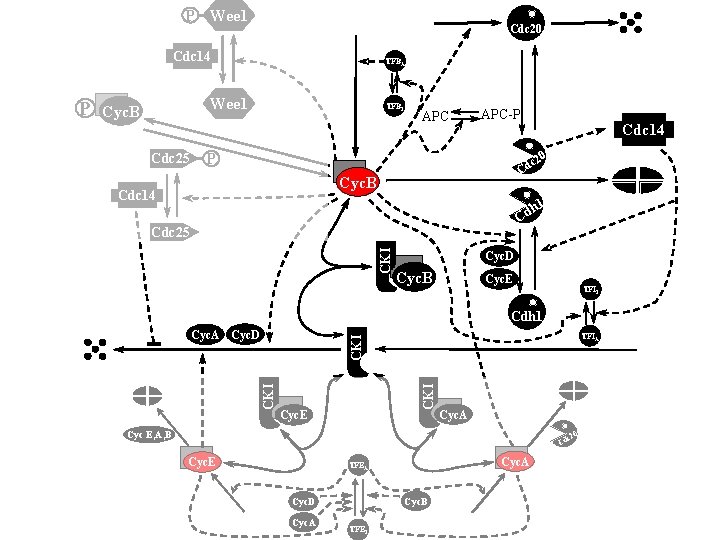
P Wee 1 Cdc 20 Cdc 14 Wee 1 Cyc. B Cdc 25 TFBI APC-P Cdc 14 0 P c 2 Cd Cyc. B Cdc 14 h 1 Cd CKI Cdc 25 Cyc. D Cyc. B Cyc. E TFII Cdh 1 TFIA CKI Cyc. A Cyc. D CKI P TFBA Cyc. E Cyc. A Cyc E, A, B 0 c 2 Cd Cyc. E Cyc. A TFEA Cyc. B Cyc. D Cyc. A TFEI

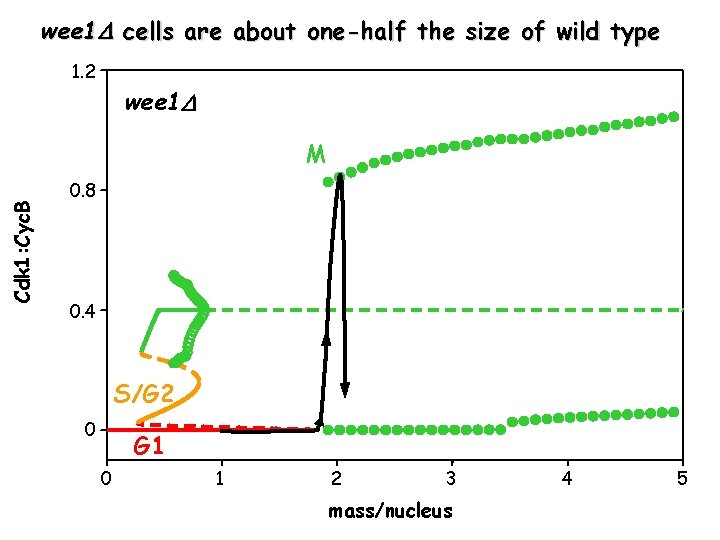
wee 1 cells are about one-half the size of wild type 1. 2 wee 1 Cdk 1: Cyc. B M 0. 8 0. 4 S/G 2 0 G 1 0 1 2 3 mass/nucleus 4 5

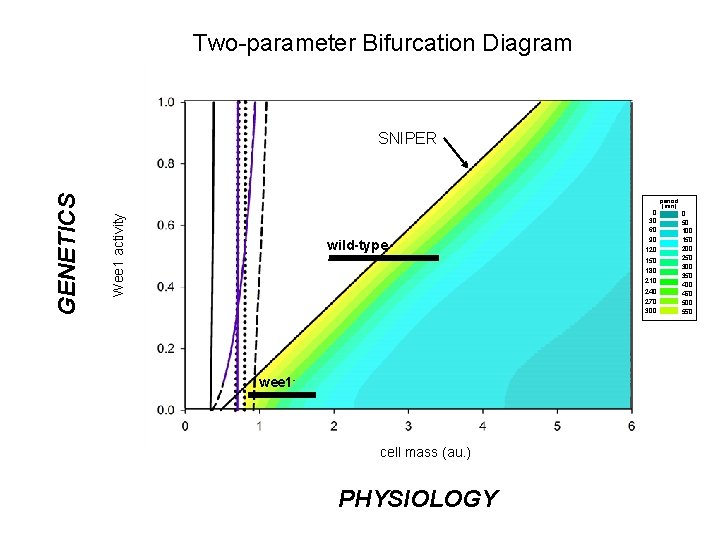
Two-parameter Bifurcation Diagram Wee 1 activity GENETICS SNIPER wild-type wee 1 - cell mass (au. ) PHYSIOLOGY 0 30 60 90 120 150 180 210 240 270 300 period (min) 0 50 100 150 200 250 300 350 400 450 500 550

P Wee 1 Cdc 20 Cdc 14 Wee 1 Cyc. B Cdc 25 TFBI APC-P Cdc 14 0 P c 2 Cd Cyc. B Cdc 14 h 1 Cd CKI Cdc 25 Cyc. D Cyc. B Cyc. E TFII Cdh 1 TFIA CKI Cyc. A Cyc. D CKI P TFBA Cyc. E Cyc. A Cyc E, A, B 0 c 2 Cd Cyc. E Cyc. A TFEA Cyc. B Cyc. D Cyc. A TFEI

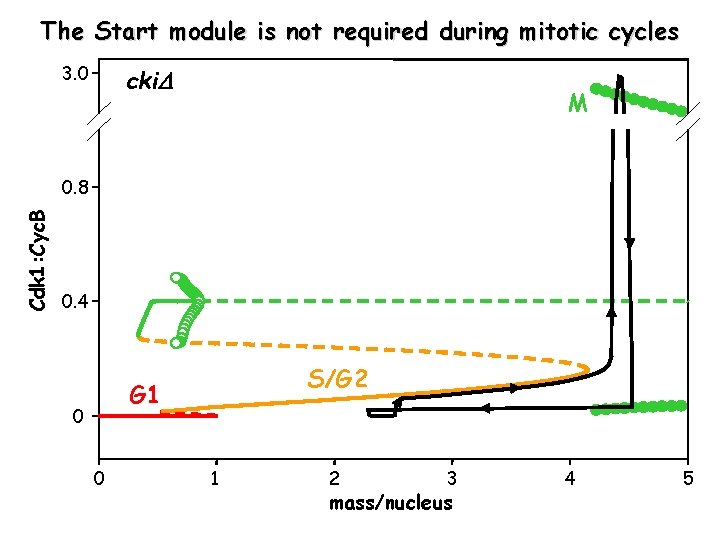
The Start module is not required during mitotic cycles 3. 0 cki M Cdk 1: Cyc. B 0. 8 0. 4 S/G 2 G 1 0 0 1 2 3 mass/nucleus 4 5

P Wee 1 Cdc 20 Cdc 14 Wee 1 Cyc. B Cdc 25 TFBI APC-P Cdc 14 0 P c 2 Cd Cyc. B Cdc 14 h 1 Cd CKI Cdc 25 Cyc. D Cyc. B Cyc. E TFII Cdh 1 TFIA CKI Cyc. A Cyc. D CKI P TFBA Cyc. E Cyc. A Cyc E, A, B 0 c 2 Cd Cyc. E Cyc. A TFEA Cyc. B Cyc. D Cyc. A TFEI

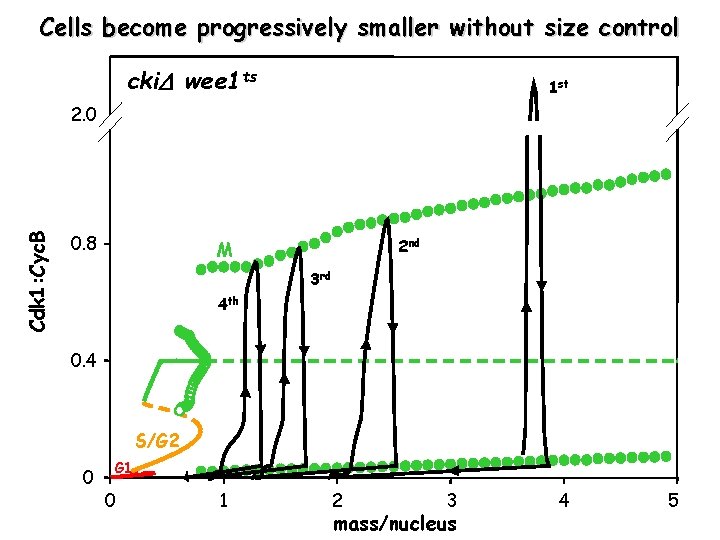
Cells become progressively smaller without size control cki wee 1 ts 1 st Cdk 1: Cyc. B 2. 0 0. 8 2 nd M 3 rd 4 th 0. 4 S/G 2 0 G 1 0 1 2 3 mass/nucleus 4 5

P Wee 1 Cdc 20 Cdc 14 Wee 1 Cyc. B Cdc 25 TFBI APC-P Cdc 14 0 P c 2 Cd Cyc. B Cdc 14 h 1 Cd CKI Cdc 25 Cyc. D Cyc. B Cyc. E TFII Cdh 1 TFIA CKI Cyc. A Cyc. D CKI P TFBA Cyc. E Cyc. A Cyc E, A, B 0 c 2 Cd Cyc. E Cyc. A TFEA Cyc. B Cyc. D Cyc. A TFEI

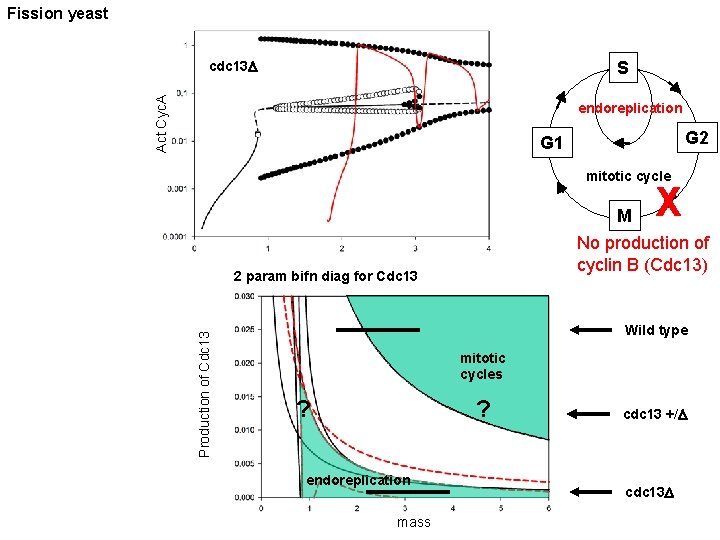
Fission yeast S Act Cyc. A cdc 13 D endoreplication G 2 G 1 mitotic cycle M No production of cyclin B (Cdc 13) 2 param bifn diag for Cdc 13 Production of Cdc 13 X Wild type mitotic cycles ? ? endoreplication mass cdc 13 +/D cdc 13 D

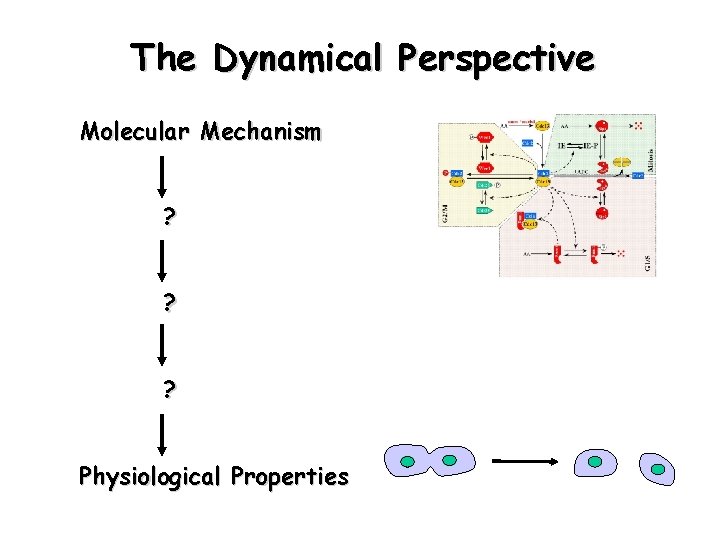
The Dynamical Perspective Molecular Mechanism ? ? ? Physiological Properties

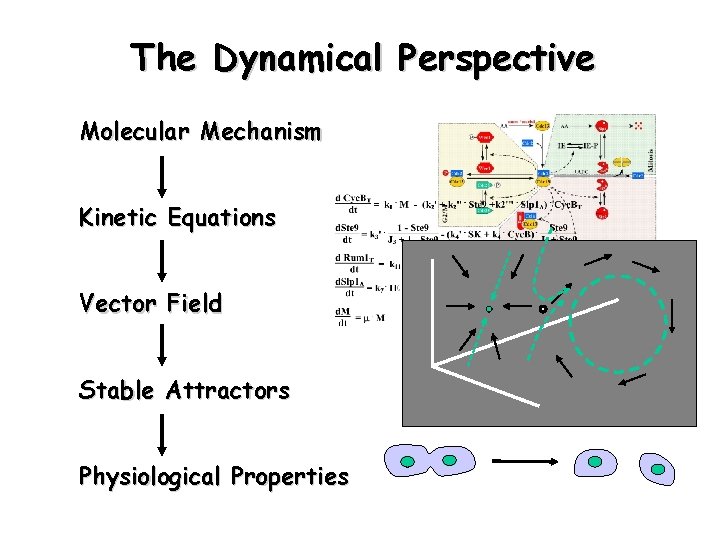
The Dynamical Perspective Molecular Mechanism Kinetic Equations Vector Field Stable Attractors Physiological Properties

References • Tyson, Chen & Novak, “Network dynamics and cell physiology, ” Nature Rev. Molec. Cell Biol. 2: 908 (2001). • Tyson, Csikasz-Nagy & Novak, “The dynamics of cell cycle regulation, ” Bio. Essays 24: 1095 (2002). • Tyson, Chen & Novak, “Sniffers, buzzers, toggles and blinkers, ” Curr. Opin. Cell Biol. 15: 221 (2003).