Macromolecular refinement with REFMAC 5 and SKETCHER of




















- Slides: 42

Macromolecular refinement with REFMAC 5 and SKETCHER of the CCP 4 suite Roberto A. Steiner – University of York

Organization 1 General aspects of refinement and overview of REFMAC 5 • • TLS Dictionary 2 Demo • • TLS refinement in REFMAC 5 SKETCHER 3 Future

1 General aspects of refinement and overview of REFMAC 5

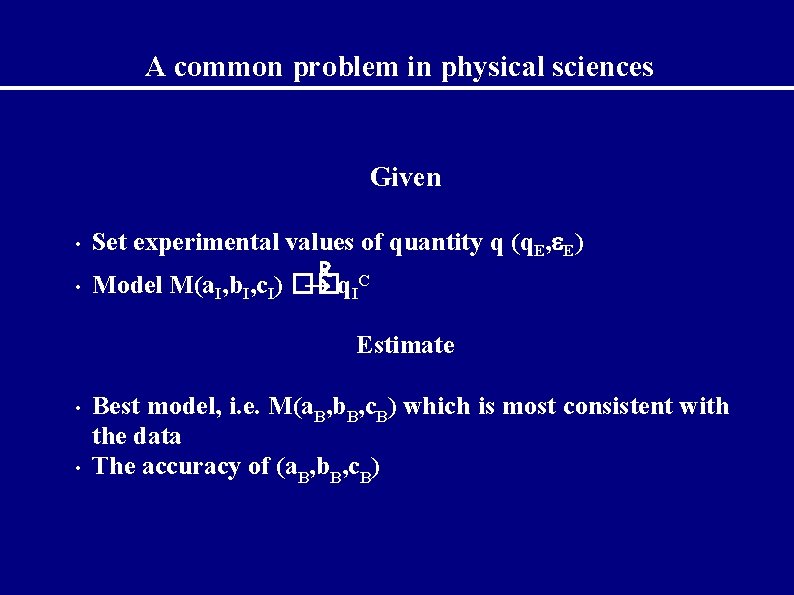
A common problem in physical sciences Given • • Set experimental values of quantity q (q. E, E) R Model M(a. I, b. I, c. I) �� q IC Estimate • • Best model, i. e. M(a. B, b. B, c. B) which is most consistent with the data The accuracy of (a. B, b. B, c. B)

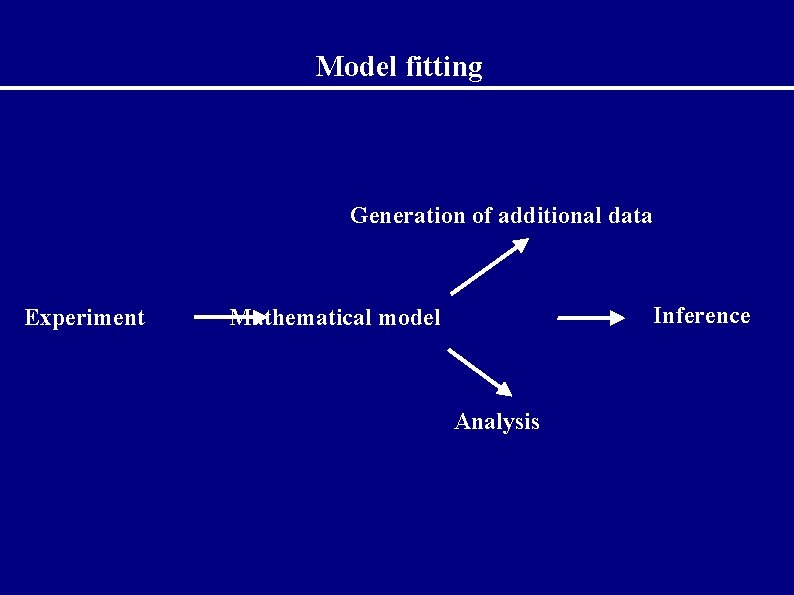
Model fitting Generation of additional data Experiment Inference Mathematical model Analysis

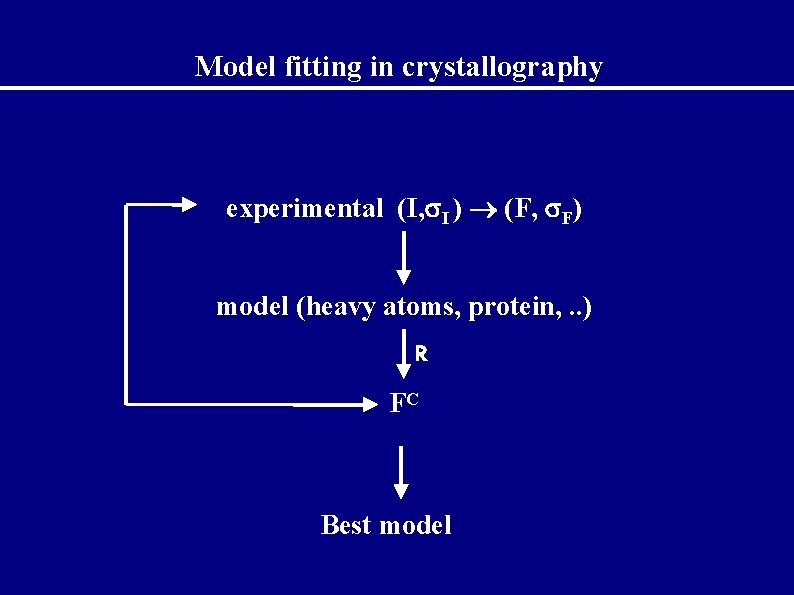
Model fitting in crystallography experimental (I, I ) (F, F) model (heavy atoms, protein, . . ) R FC Best model

Key aspects in model fitting • Parameterization of the model • Type of residual • Type of minimization • Prior information

Bayesian approach The best model is the one which has highest probability given a set of observations and a certain prior knowledge. BAYES' THEOREM P(M; O)=P(M)P(O; M)/P(O) Probability Theory: The Logic of Science by E. T. Jaynes http: //bayes. wustl. edu

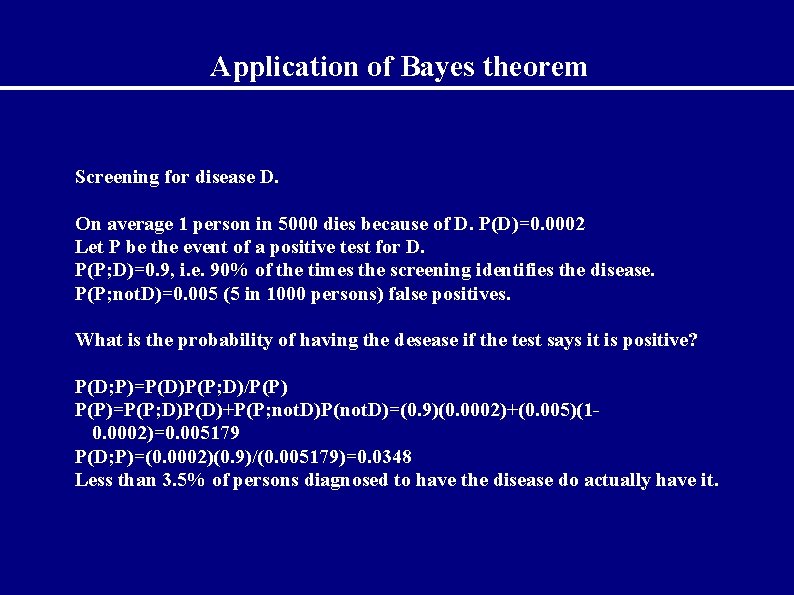
Application of Bayes theorem Screening for disease D. On average 1 person in 5000 dies because of D. P(D)=0. 0002 Let P be the event of a positive test for D. P(P; D)=0. 9, i. e. 90% of the times the screening identifies the disease. P(P; not. D)=0. 005 (5 in 1000 persons) false positives. What is the probability of having the desease if the test says it is positive? P(D; P)=P(D)P(P; D)/P(P)=P(P; D)P(D)+P(P; not. D)P(not. D)=(0. 9)(0. 0002)+(0. 005)(10. 0002)=0. 005179 P(D; P)=(0. 0002)(0. 9)/(0. 005179)=0. 0348 Less than 3. 5% of persons diagnosed to have the disease do actually have it.

Maximum likelihood residual P(M; O) = P(M)P(O; M)/P(O) = P(M)L(M; O) max P(M; O) min [-log. P(M) -log. L(M; O)] Murshudov et al. , Acta Cryst. (1997) D 53, 240 -255

Maximum likelihood refinement programs • REFMAC 5 • CNS/CNX • BUSTER-TNT

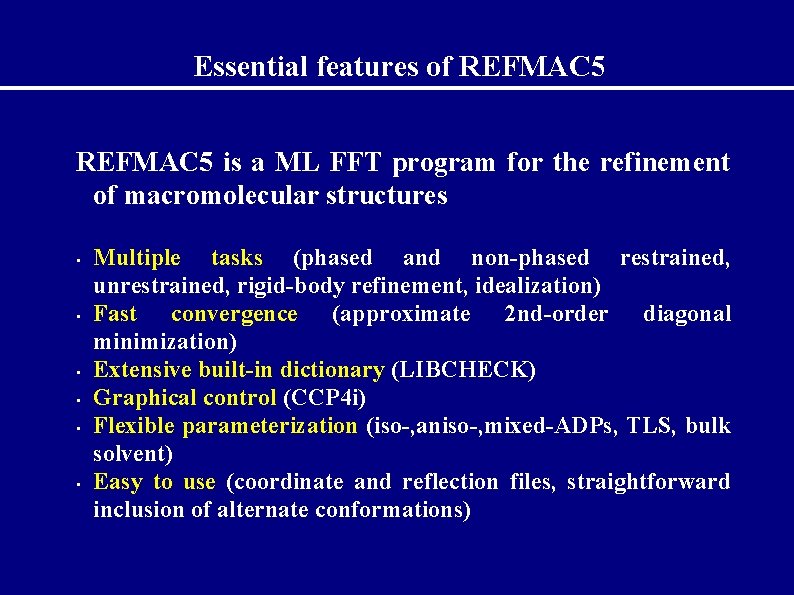
Essential features of REFMAC 5 is a ML FFT program for the refinement of macromolecular structures • • • Multiple tasks (phased and non-phased restrained, unrestrained, rigid-body refinement, idealization) Fast convergence (approximate 2 nd-order diagonal minimization) Extensive built-in dictionary (LIBCHECK) Graphical control (CCP 4 i) Flexible parameterization (iso-, aniso-, mixed-ADPs, TLS, bulk solvent) Easy to use (coordinate and reflection files, straightforward inclusion of alternate conformations)

Selected topic 1: TLS ADPs are an important component of a macromolecule • Proper parameterization • Biological significance Displacements are likely anisotropic, but rarely we have the luxury of refinining individual aniso-U. Instead iso-B are used. TLS parameterization allows an intermediate description.

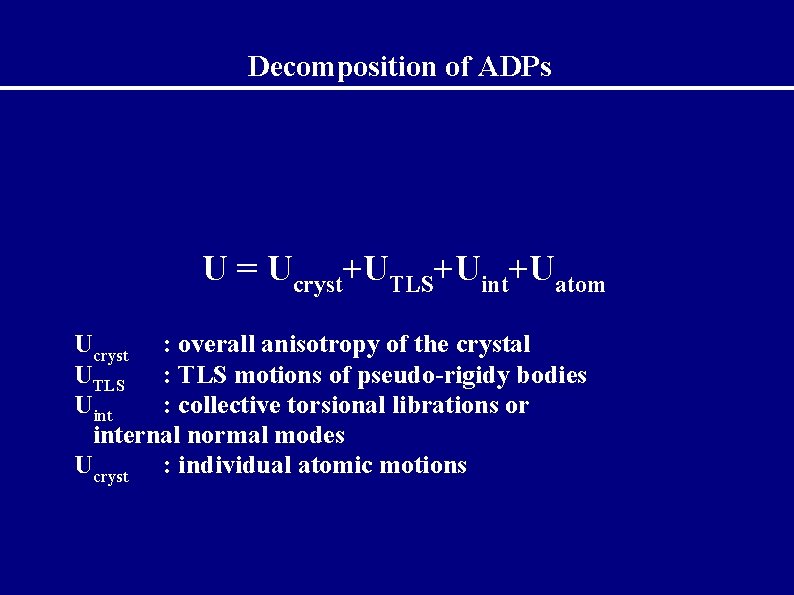
Decomposition of ADPs U = Ucryst+UTLS+Uint+Uatom Ucryst : overall anisotropy of the crystal UTLS : TLS motions of pseudo-rigidy bodies Uint : collective torsional librations or internal normal modes Ucryst : individual atomic motions

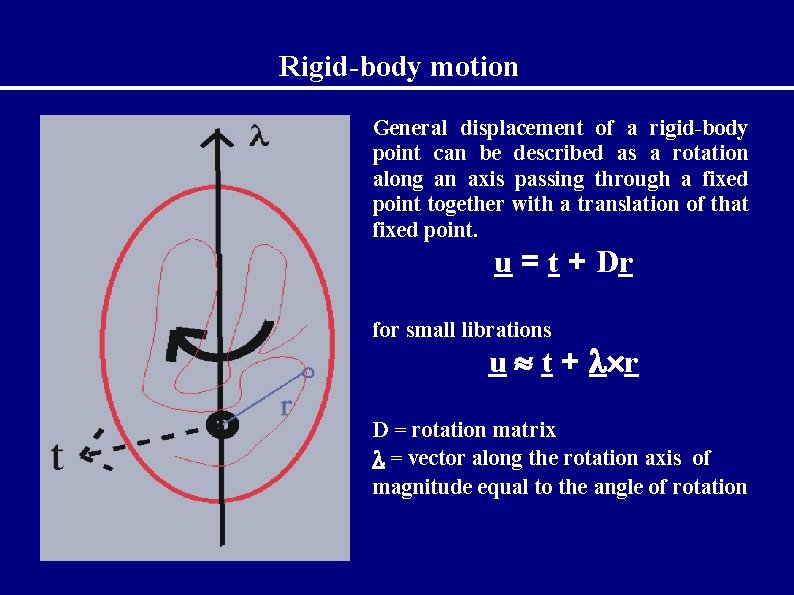
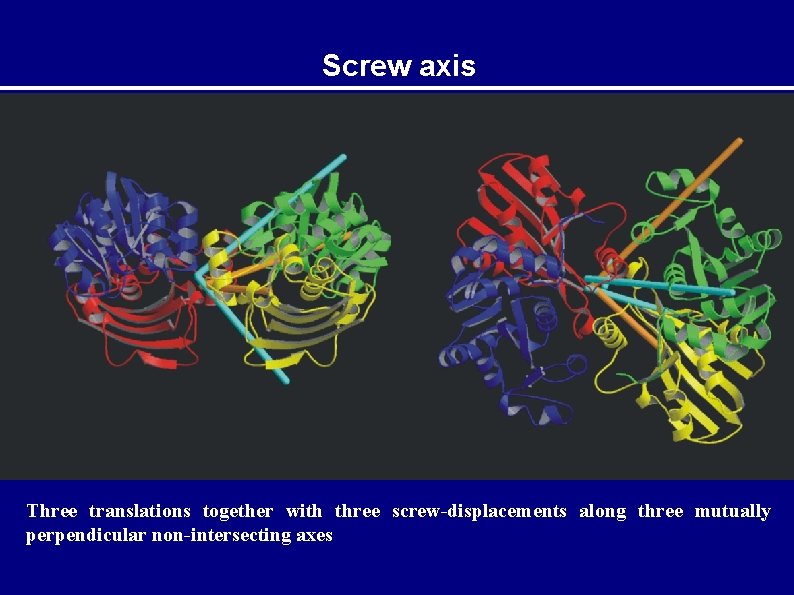
Rigid-body motion General displacement of a rigid-body point can be described as a rotation along an axis passing through a fixed point together with a translation of that fixed point. u = t + Dr for small librations u t + r D = rotation matrix = vector along the rotation axis of magnitude equal to the angle of rotation

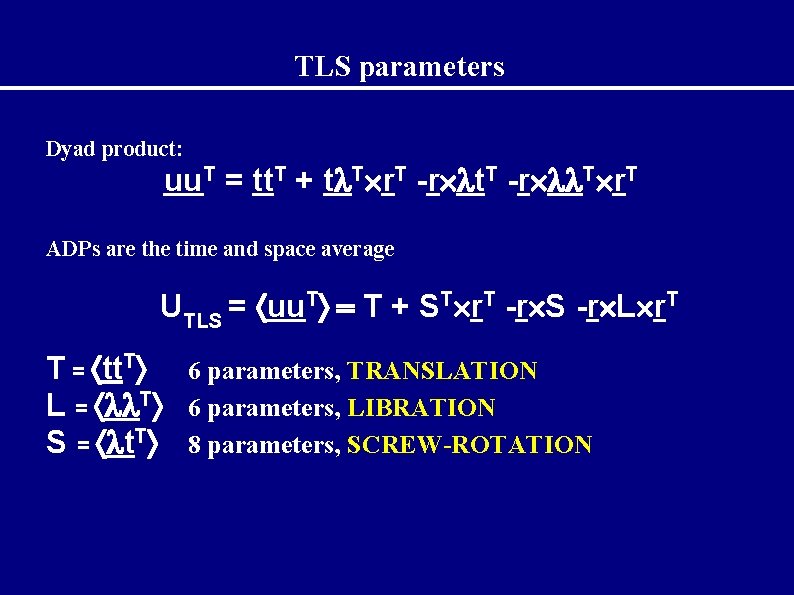
TLS parameters Dyad product: uu. T = tt. T + t T r. T -r t. T -r T r. T ADPs are the time and space average UTLS = uu. T T + ST r. T -r S -r L r. T T = tt. T 6 parameters, TRANSLATION L = T 6 parameters, LIBRATION S = t. T 8 parameters, SCREW-ROTATION

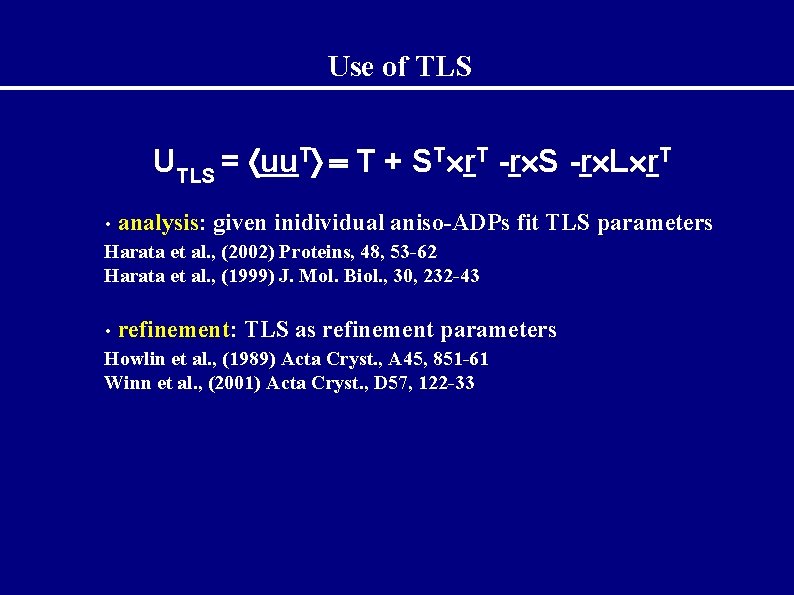
Use of TLS UTLS = uu. T T + ST r. T -r S -r L r. T • analysis: given inidividual aniso-ADPs fit TLS parameters Harata et al. , (2002) Proteins, 48, 53 -62 Harata et al. , (1999) J. Mol. Biol. , 30, 232 -43 • refinement: TLS as refinement parameters Howlin et al. , (1989) Acta Cryst. , A 45, 851 -61 Winn et al. , (2001) Acta Cryst. , D 57, 122 -33

Choice of TLS groups and resolution Choice: chains, domains, secondary structure elements, . . more complex MD, . . . Resolution: you have only 20 more parameters per TLS group. Thioredoxin reductase 3 Å (Sandalova et al. , (2001) PNAS, 98, 9533 -8) 6 TLS groups (1 for each of 6 monomers in asu)

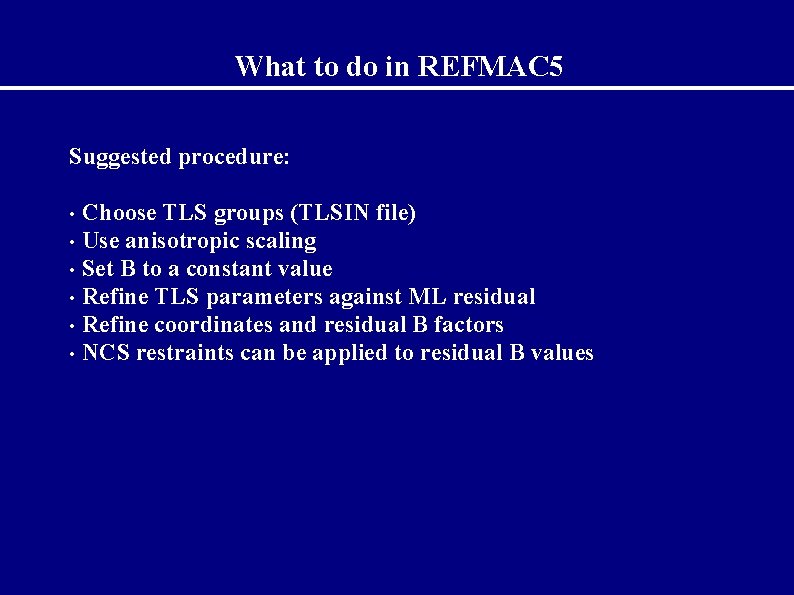
What to do in REFMAC 5 Suggested procedure: Choose TLS groups (TLSIN file) • Use anisotropic scaling • Set B to a constant value • Refine TLS parameters against ML residual • Refine coordinates and residual B factors • NCS restraints can be applied to residual B values •

What to do with output Check Rfree and TLS parameters for convergence • Check TLS parameters to see if there is any dominant displacement • Pass XYZOUT and TLSOUT through TLSANL for analysis •

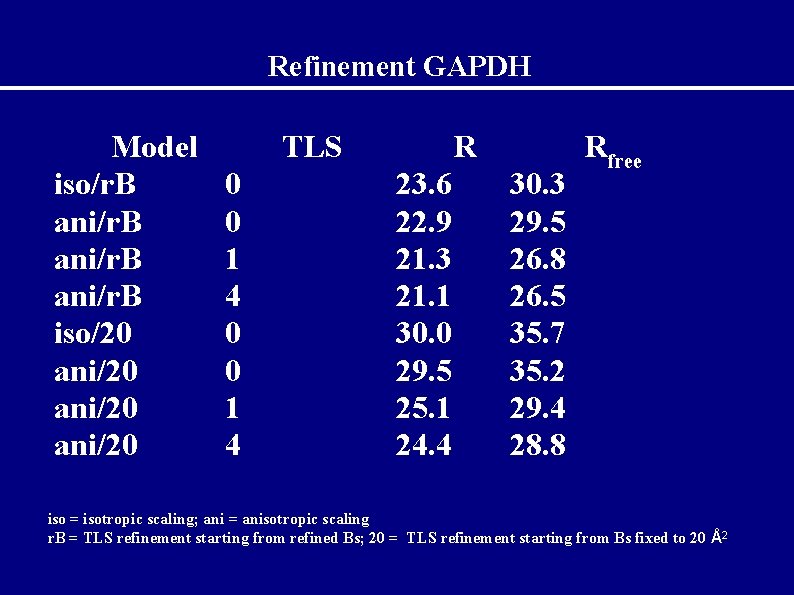
Example GAPDH Glyceraldehyde-3 -phosphate dehydrogenase from Sulfolobus solfataricus (Isupov et al. , (1999) J. Mol. Biol. , 291, 651● 60) 340 amino acids ● 2 chains in asymmetric unit (O and Q), each molecule has NAD-binding and catalytic domains. ● P 41212, data to 2. 05Å ●

GAPDH before and after TLS R 0 22. 9 29. 5 1 4 4/NCS 21. 4 21. 1 22. 0 25. 9 25. 8 25. 7 Rfree

Refinement GAPDH Model iso/r. B ani/r. B iso/20 ani/20 TLS 0 0 1 4 R 23. 6 22. 9 21. 3 21. 1 30. 0 29. 5 25. 1 24. 4 30. 3 29. 5 26. 8 26. 5 35. 7 35. 2 29. 4 28. 8 Rfree iso = isotropic scaling; ani = anisotropic scaling r. B = TLS refinement starting from refined Bs; 20 = TLS refinement starting from Bs fixed to 20 Å2

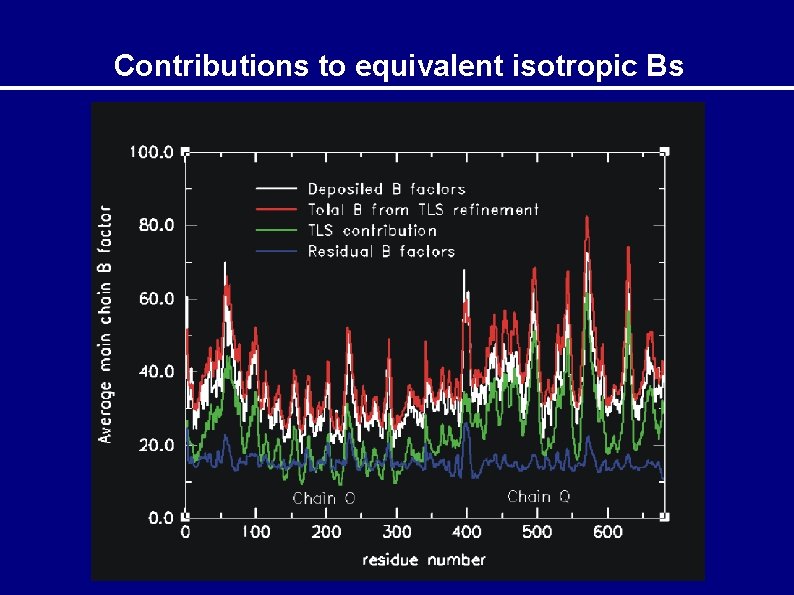
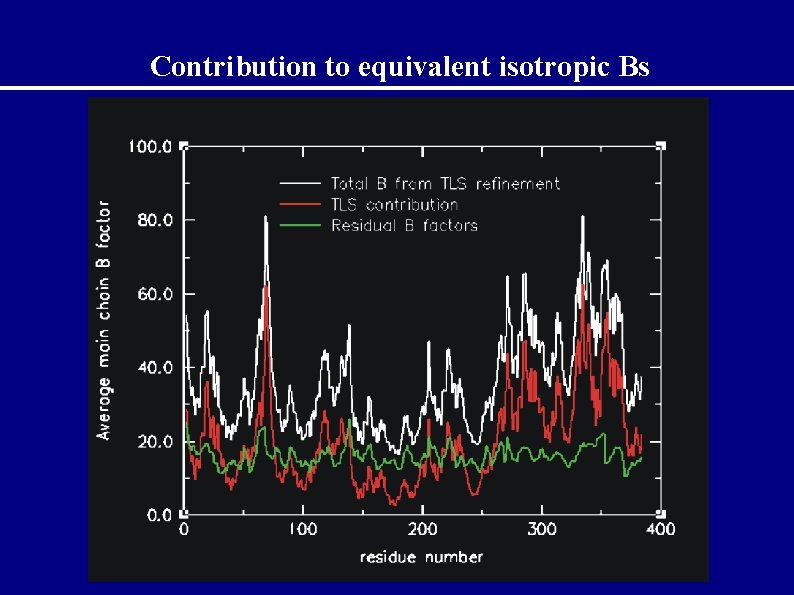
Contributions to equivalent isotropic Bs

Screw axis Three translations together with three screw-displacements along three mutually perpendicular non-intersecting axes

Example Ger. E Transcription regulator from Bacillus subtilis (Ducrois et al. , (2001) J. Mol. Biol. , 306, 759 -71). ● 74 amino acids ● Six chains A-F in asymmetric unit ● C 2, data to 2. 05Å ●

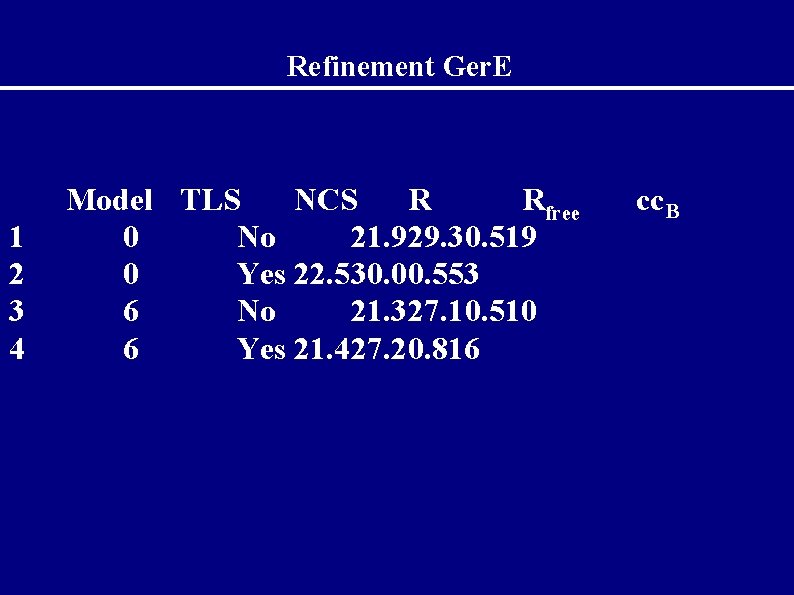
Refinement Ger. E 1 2 3 4 Model TLS NCS R Rfree 0 No 21. 929. 30. 519 0 Yes 22. 530. 00. 553 6 No 21. 327. 10. 510 6 Yes 21. 427. 20. 816 cc. B

Contribution to equivalent isotropic Bs

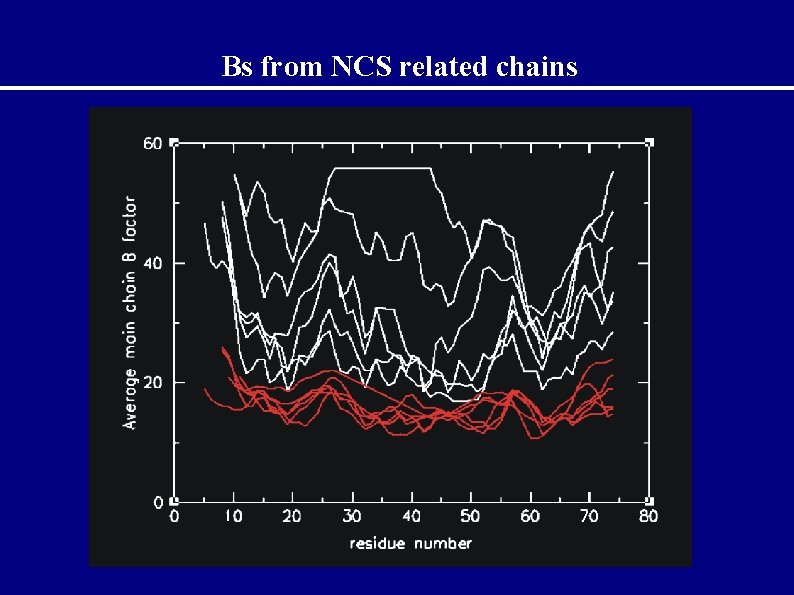
Bs from NCS related chains

Summary TLS parameterization allows to partly take into account anisotropic motions at modest resolution (> 3. 5 Å) • TLS refinement might improve refinement statistics of several percent • TLS refinement in REFMAC 5 is fast and therefore can be used routinely •

Selected topic 2: dictionary The use of prior knowledge requires its organized storage. $CCP 4/html/mon_lib. html www. ysbl. york. ac. uk/~alexei/dictionary. html

Monomer description REFMAC 5 requires a complete chemical description of all monomers (any molecular entity) present in the input coordinate file About 2000 common monomers are already ($CLIBD_MON = $CCP 4/lib/data/monomers) • • Monomer and atoms identifier Element symbol Energy type Partial charge Covalent bonds (target values and SDs) Torsion angles (target values and SDs) Chiral centers Planes present

Monomer library $CCP 4/lib/data/monomers/ ener_lib. cif mon_lib_com. cif modifications mon_lib_list. html 0/, 1/, . . . -definition of atom types -definition of links and -missing file in version 4. 2 -definition of various monomers

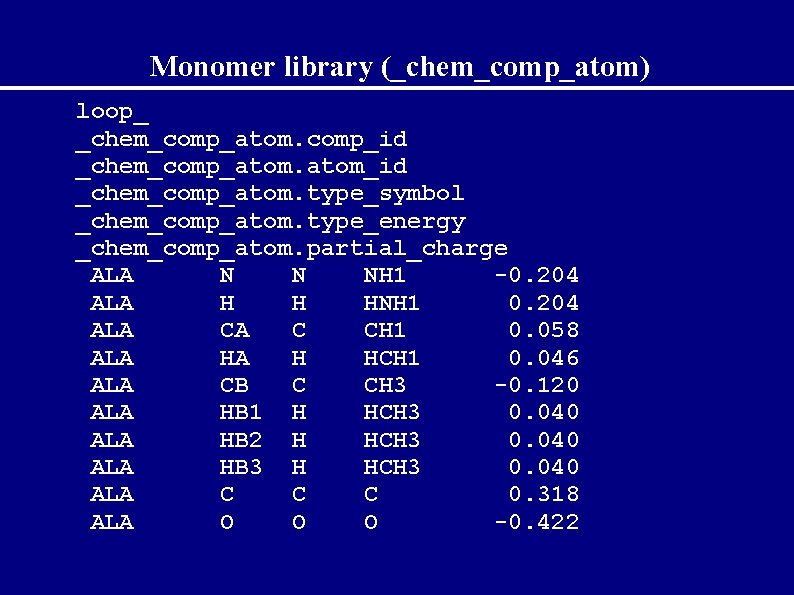
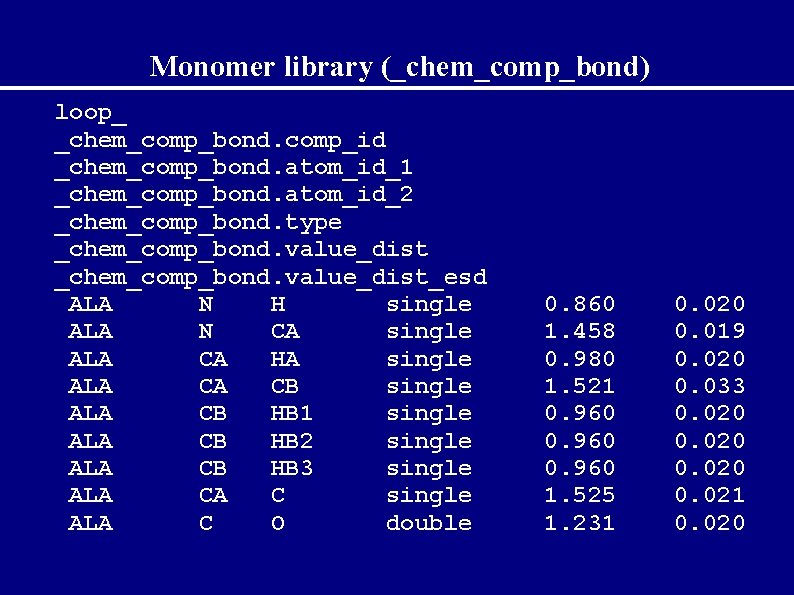
Description of monomers In the files: */###. cif For every monomer contain catagories: _chem_comp_atom _chem_comp_bond _chem_comp_angle _chem_comp_tor _chem_comp_chir _chem_comp_plane_atom

Monomer library (_chem_comp_atom) loop_ _chem_comp_atom. comp_id _chem_comp_atom_id _chem_comp_atom. type_symbol _chem_comp_atom. type_energy _chem_comp_atom. partial_charge ALA N N NH 1 -0. 204 ALA H H HNH 1 0. 204 ALA CA C CH 1 0. 058 ALA HA H HCH 1 0. 046 ALA CB C CH 3 -0. 120 ALA HB 1 H HCH 3 0. 040 ALA HB 2 H HCH 3 0. 040 ALA HB 3 H HCH 3 0. 040 ALA C C C 0. 318 ALA O O O -0. 422

Monomer library (_chem_comp_bond) loop_ _chem_comp_bond. comp_id _chem_comp_bond. atom_id_1 _chem_comp_bond. atom_id_2 _chem_comp_bond. type _chem_comp_bond. value_dist_esd ALA N H single ALA N CA single ALA CA HA single ALA CA CB single ALA CB HB 1 single ALA CB HB 2 single ALA CB HB 3 single ALA CA C single ALA C O double 0. 860 1. 458 0. 980 1. 521 0. 960 1. 525 1. 231 0. 020 0. 019 0. 020 0. 033 0. 020 0. 021 0. 020

What happens when you run REFMAC 5 • • You have a monomer for which there is a complete description The program carries on and takes everything from the dictionary You have a monomer for which there is only a minimal description or no description The program tries to generate a complete library description and then STOPS for you to check it.

SKETCHER If a monomer is not in the library then SKETCHER can be used SKETCHER is a graphical interface to LIBCHECK which creates new monomer library description

IT A W O I O N T O E T N AV A H N L A 'L S N YOU I T ID N O O I N Demo A S E FR A CC E W M O ' I U T ) O 002 EX Y IF A 2 E N C A ( R TH FO 2

3 Future (near and far)

Future • Fast calculation of complete Hessian matrix • Refinement along internal degrees of freedom • Refinement using anomalous data • Bayesian refinement of twinned data • Lots more

People • Garib N. Murshudov, York • Alexei Vaguine, York • Martyn Winn*, CCP 4 • Liz Potterton*, York • Eleanor Dodson, York • Kim Hendrik, EBI Cambridge • people who gave their data * kindly provided many of the slides presented here Financial support • CCP 4 • Wellcome Trust