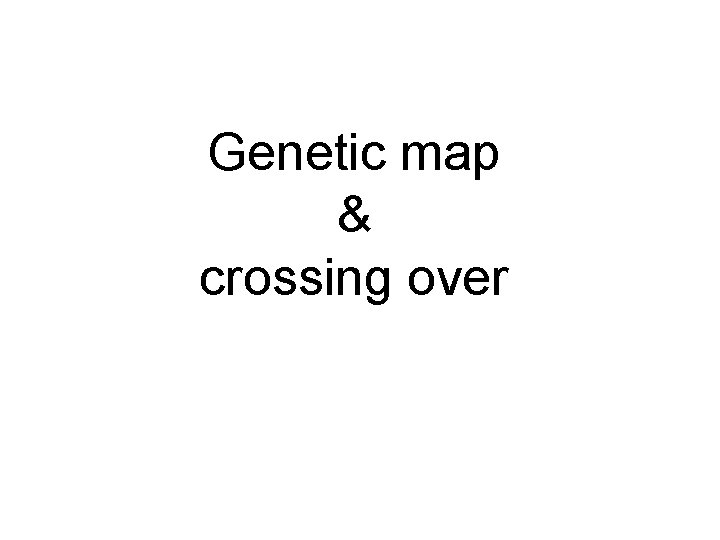
Genetic map crossing over Linked assortment Rr Yy






- Slides: 26

Genetic map & crossing over

Linked assortment Rr. Yy ry RY Possible gametes. IS NOT ALWAYS TRUE

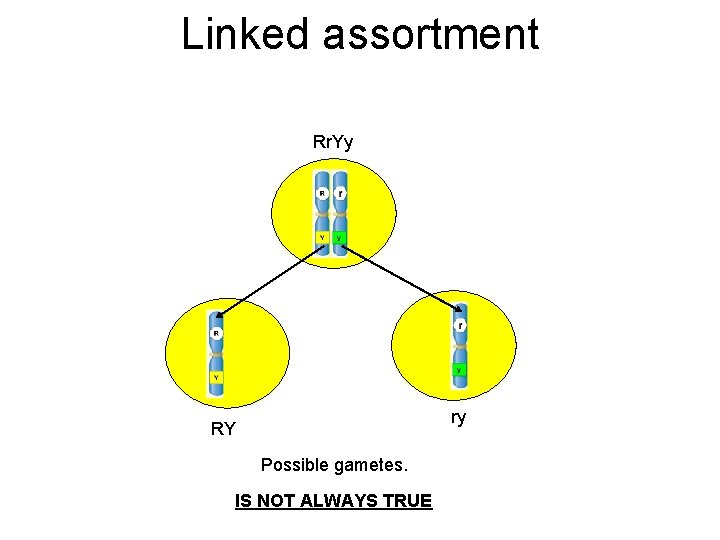
Morgan Provided Evidence for the Linkage of Several X-linked Genes • The first direct evidence of linkage came from studies of Thomas Hunt Morgan • Morgan investigated several traits that followed an X-linked pattern of inheritance – Body color – Eye color – Wing length

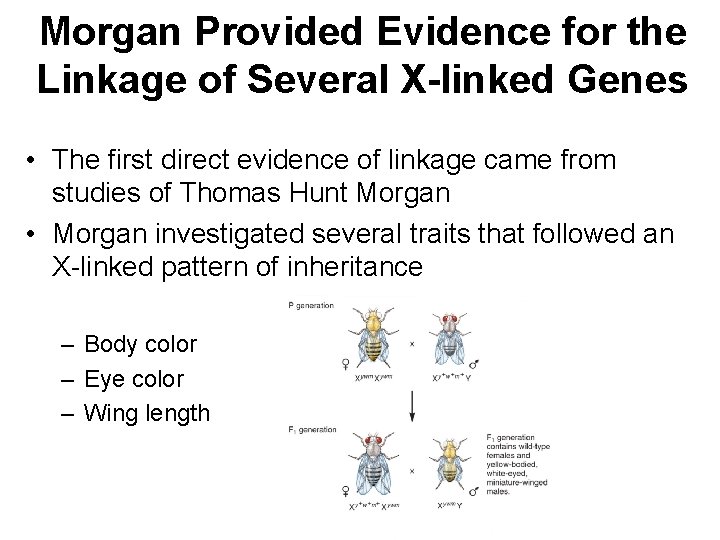
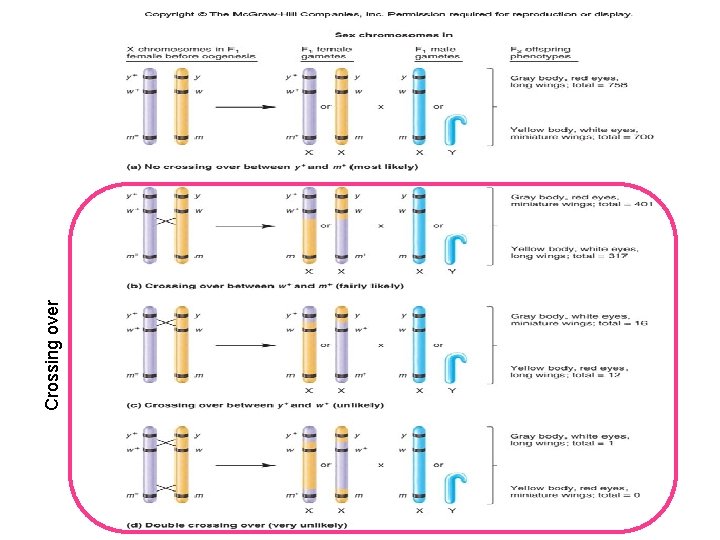
P Males P Females • Morgan observed a much higher proportion of the combinations of traits found in the parental generation • Morgan’s explanation: – All three genes are located on the X chromosome – Therefore, they tend to be transmitted together as a unit

Morgan Provided Evidence for the Linkage of Several X-linked Genes • However, Morgan still had to interpret two key observations – 1. Why did the F 2 generation have a significant number of non parental combinations? – 2. Why was there a quantitative difference between the various non parental combinations?

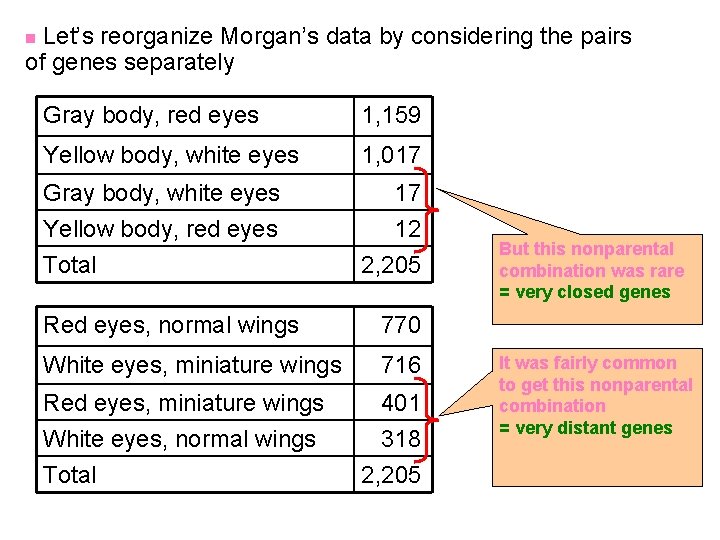
Let’s reorganize Morgan’s data by considering the pairs of genes separately n Gray body, red eyes 1, 159 Yellow body, white eyes 1, 017 Gray body, white eyes Yellow body, red eyes Total 17 12 2, 205 Red eyes, normal wings 770 White eyes, miniature wings 716 Red eyes, miniature wings White eyes, normal wings 401 318 Total 2, 205 But this nonparental combination was rare = very closed genes It was fairly common to get this nonparental combination = very distant genes

• Morgan made three important hypotheses to explain his results – 1. The genes for body color, eye color and wing length are all located on the X-chromosome • They tend to be inherited together – 2. Due to crossing over, the homologous X chromosomes (in the female) can exchange pieces of chromosomes • This created new combination of alleles – 3. The likelihood of crossing over depends on the distance between the two genes • Crossing over is more likely to occur between two genes that are far apart from each other

Crossing over

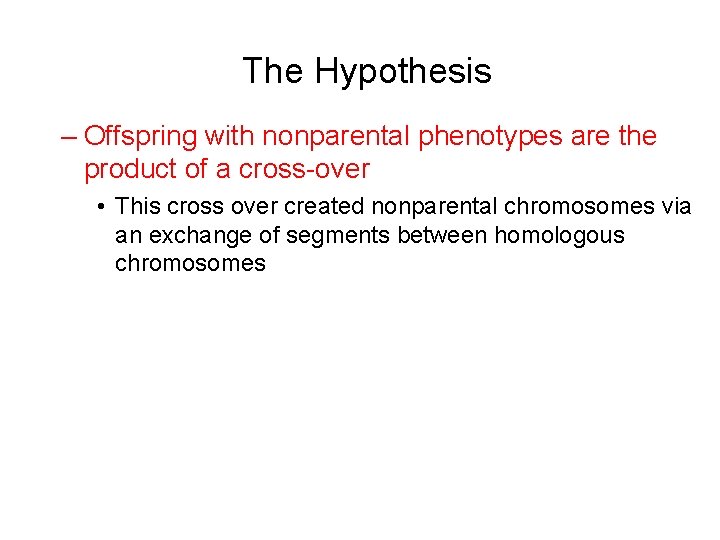
The Hypothesis – Offspring with nonparental phenotypes are the product of a cross-over • This cross over created nonparental chromosomes via an exchange of segments between homologous chromosomes

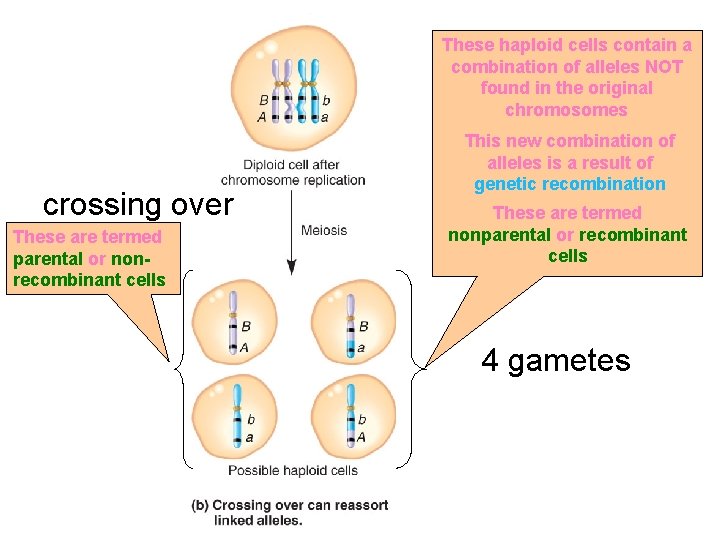
The haploid cells contain the same combination of alleles as the original chromosomes The arrangement of linked alleles has not been altered No crossing over 2 gametes

These haploid cells contain a combination of alleles NOT found in the original chromosomes crossing over These are termed parental or nonrecombinant cells This new combination of alleles is a result of genetic recombination These are termed nonparental or recombinant cells 4 gametes

GENETIC MAPPING • Genetic mapping is also known as gene mapping, chromosome mapping or linkage analysis • Its purpose is to determine the linear order of linked genes along the same chromosome

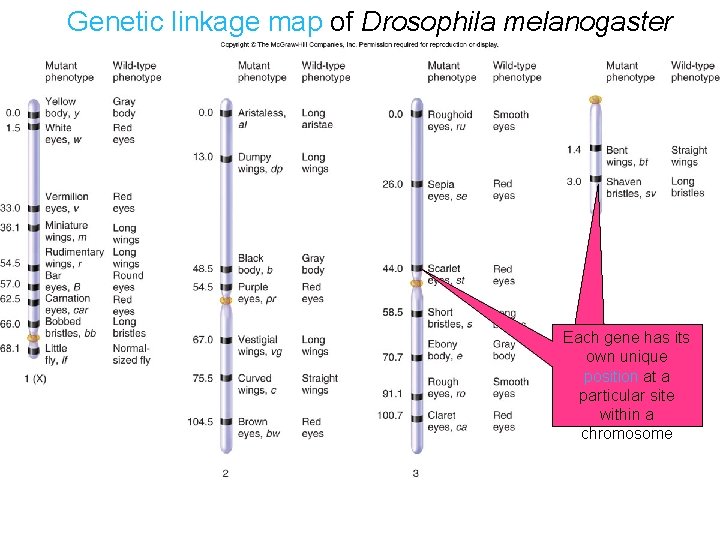
Genetic linkage map of Drosophila melanogaster Each gene has its own unique position at a particular site within a chromosome

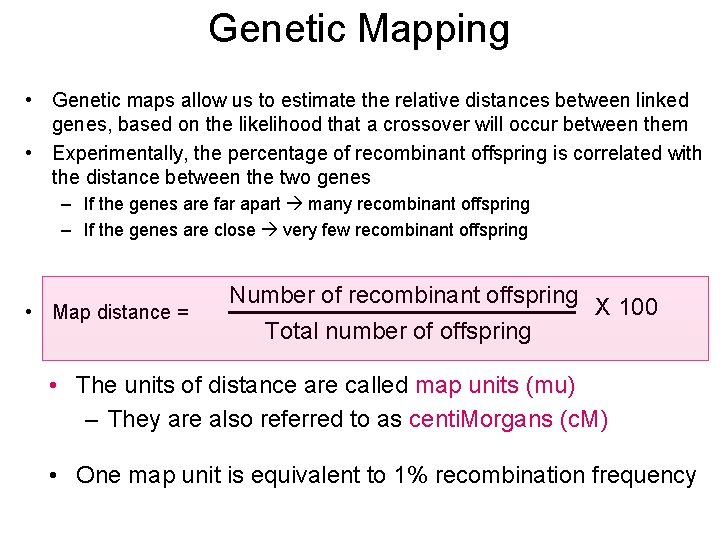
Genetic Mapping • Genetic maps allow us to estimate the relative distances between linked genes, based on the likelihood that a crossover will occur between them • Experimentally, the percentage of recombinant offspring is correlated with the distance between the two genes – If the genes are far apart many recombinant offspring – If the genes are close very few recombinant offspring • Map distance = Number of recombinant offspring X 100 Total number of offspring • The units of distance are called map units (mu) – They are also referred to as centi. Morgans (c. M) • One map unit is equivalent to 1% recombination frequency

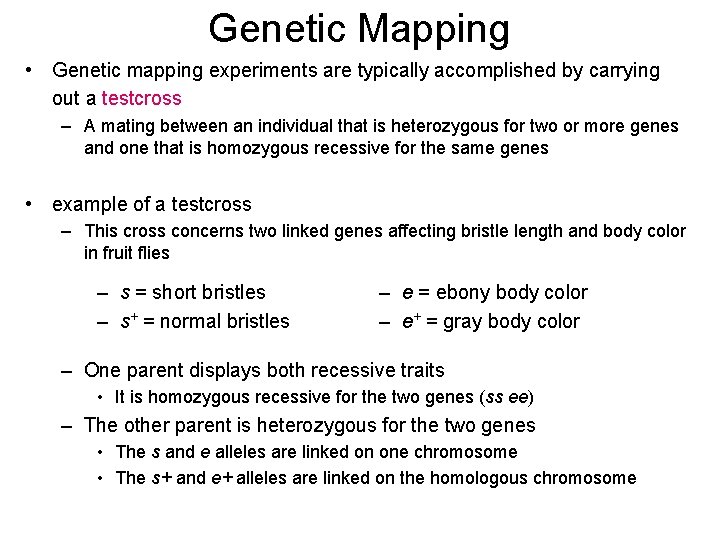
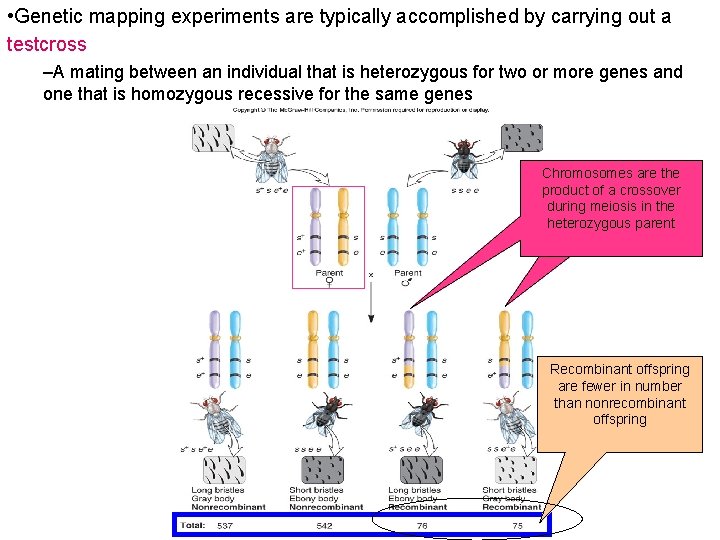
Genetic Mapping • Genetic mapping experiments are typically accomplished by carrying out a testcross – A mating between an individual that is heterozygous for two or more genes and one that is homozygous recessive for the same genes • example of a testcross – This cross concerns two linked genes affecting bristle length and body color in fruit flies – s = short bristles – s+ = normal bristles – e = ebony body color – e+ = gray body color – One parent displays both recessive traits • It is homozygous recessive for the two genes (ss ee) – The other parent is heterozygous for the two genes • The s and e alleles are linked on one chromosome • The s+ and e+ alleles are linked on the homologous chromosome

• Genetic mapping experiments are typically accomplished by carrying out a testcross –A mating between an individual that is heterozygous for two or more genes and one that is homozygous recessive for the same genes Chromosomes are the product of a crossover during meiosis in the heterozygous parent Recombinant offspring are fewer in number than nonrecombinant offspring

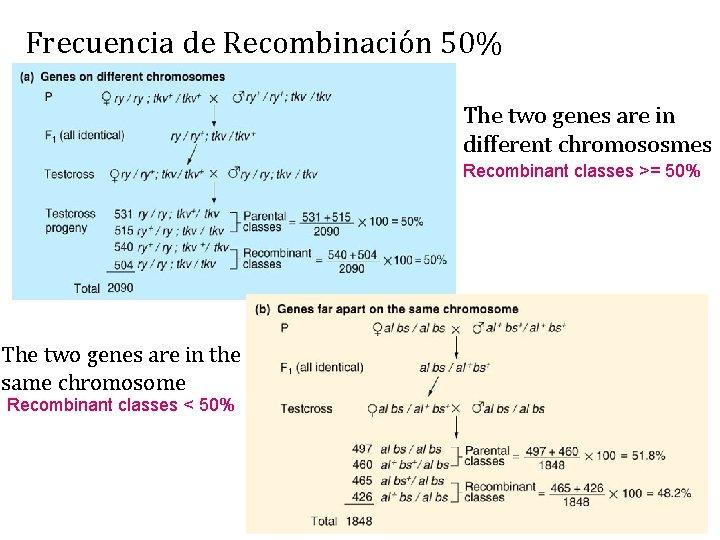
Frecuencia de Recombinación 50% The two genes are in different chromososmes Recombinant classes >= 50% The two genes are in the same chromosome Recombinant classes < 50%

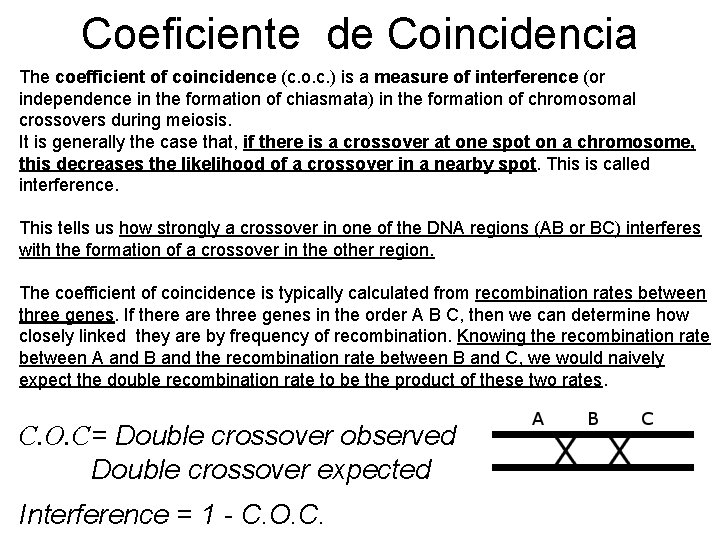
Coeficiente de Coincidencia The coefficient of coincidence (c. o. c. ) is a measure of interference (or independence in the formation of chiasmata) in the formation of chromosomal crossovers during meiosis. It is generally the case that, if there is a crossover at one spot on a chromosome, this decreases the likelihood of a crossover in a nearby spot. This is called interference. This tells us how strongly a crossover in one of the DNA regions (AB or BC) interferes with the formation of a crossover in the other region. The coefficient of coincidence is typically calculated from recombination rates between three genes. If there are three genes in the order A B C, then we can determine how closely linked they are by frequency of recombination. Knowing the recombination rate between A and B and the recombination rate between B and C, we would naively expect the double recombination rate to be the product of these two rates. C. O. C= Double crossover observed Double crossover expected Interference = 1 - C. O. C.

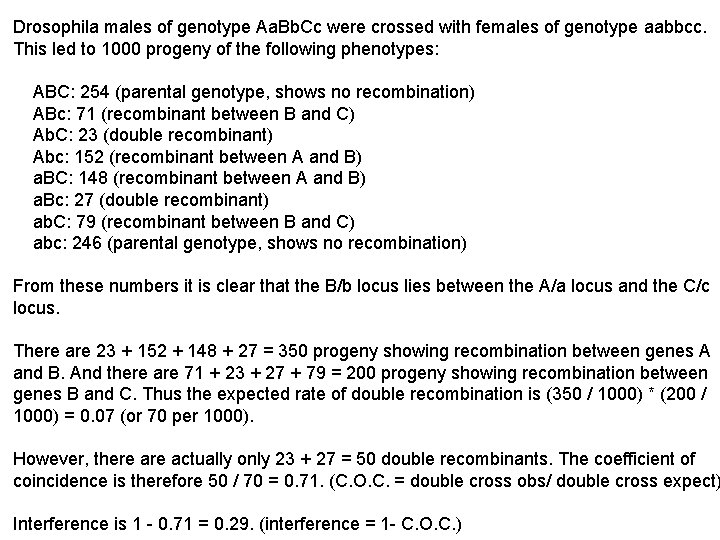
Drosophila males of genotype Aa. Bb. Cc were crossed with females of genotype aabbcc. This led to 1000 progeny of the following phenotypes: ABC: 254 (parental genotype, shows no recombination) ABc: 71 (recombinant between B and C) Ab. C: 23 (double recombinant) Abc: 152 (recombinant between A and B) a. BC: 148 (recombinant between A and B) a. Bc: 27 (double recombinant) ab. C: 79 (recombinant between B and C) abc: 246 (parental genotype, shows no recombination) From these numbers it is clear that the B/b locus lies between the A/a locus and the C/c locus. There are 23 + 152 + 148 + 27 = 350 progeny showing recombination between genes A and B. And there are 71 + 23 + 27 + 79 = 200 progeny showing recombination between genes B and C. Thus the expected rate of double recombination is (350 / 1000) * (200 / 1000) = 0. 07 (or 70 per 1000). However, there actually only 23 + 27 = 50 double recombinants. The coefficient of coincidence is therefore 50 / 70 = 0. 71. (C. O. C. = double cross obs/ double cross expect) Interference is 1 - 0. 71 = 0. 29. (interference = 1 - C. O. C. )

Intereference = It is generally the case that, if there is a crossover at one spot on a chromosome, this decreases the likelihood of a crossover in a nearby spot C. O. C. = 0, complete interference 0<C. O. C. <1 more the two genes are distant less is the interference C. OC. = 1, no interference (esperados=observados)*

v. Remember that in the Drosophila male you do not have recombination (Morgan, 1912, 1914). • Other scientific studies showed that you can actually have some really low in frequency spontaneous recombination in male (Henderson, 1977).

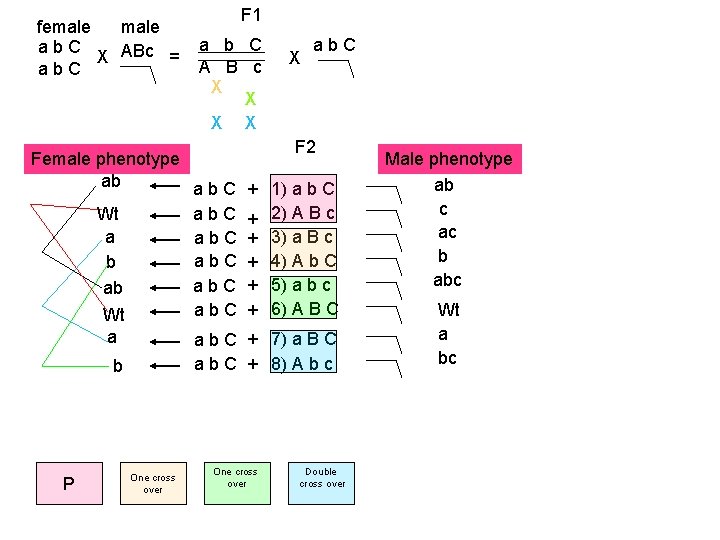
female ab. C X ABc = ab. C F 1 a b C A B c X X Female phenotype ab ab. C Wt ab. C b ab. C ab ab. C Wt a F 2 + 1) a b C + 2) A B c + 3) a B c + 4) A b C + 5) a b c + 6) A B C a b C + 7) a B C a b C + 8) A b c b P X ab. C One cross over Double cross over Male phenotype ab c ac b abc Wt a bc

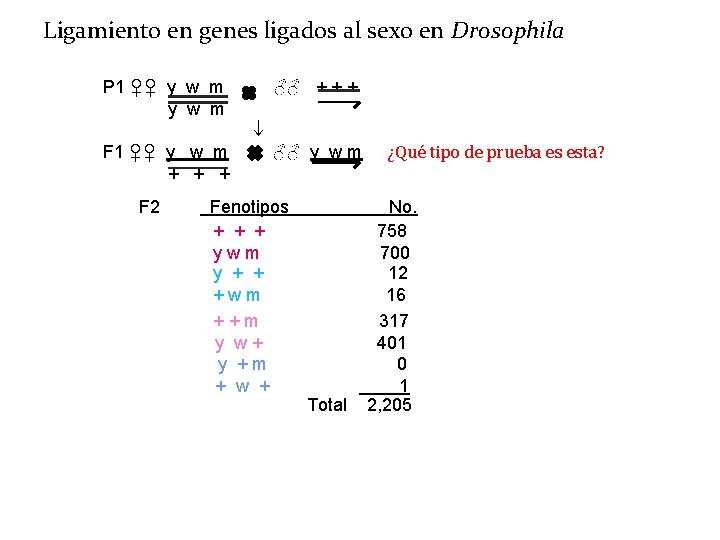
Ligamiento en genes ligados al sexo en Drosophila P 1 ♀♀ y w m F 1 ♀♀ y w m + + + F 2 NCO COS-1 COS-2 COD ♂♂ + + + ♂♂ y w m Fenotipos + + + ywm y + + +wm ++m y w+ y +m + w + Total ¿Qué tipo de prueba es esta? No. 758 700 12 16 317 401 0 1 2, 205

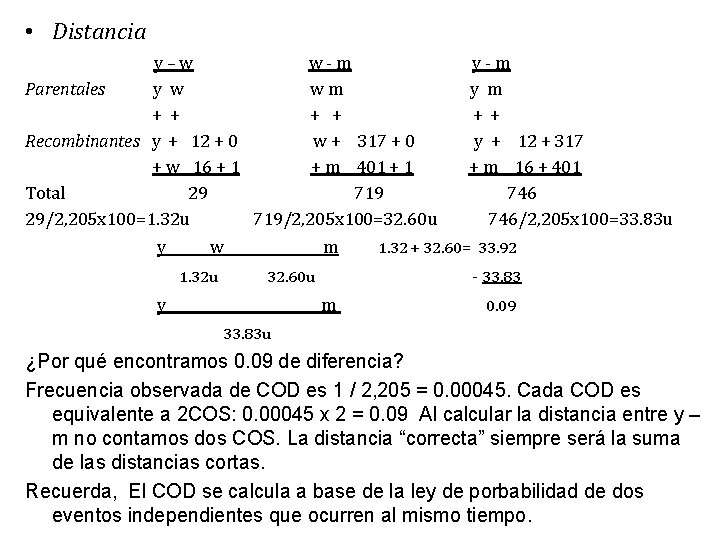
• Distancia y–w w-m Parentales y w wm + + Recombinantes y + 12 + 0 w + 317 + 0 + w 16 + 1 + m 401 + 1 Total 29 719 29/2, 205 x 100=1. 32 u 719/2, 205 x 100=32. 60 u y w 1. 32 u m 32. 60 u y y-m y m + + y + 12 + 317 + m 16 + 401 746/2, 205 x 100=33. 83 u 1. 32 + 32. 60= 33. 92 - 33. 83 m 0. 09 33. 83 u ¿Por qué encontramos 0. 09 de diferencia? Frecuencia observada de COD es 1 / 2, 205 = 0. 00045. Cada COD es equivalente a 2 COS: 0. 00045 x 2 = 0. 09 Al calcular la distancia entre y – m no contamos dos COS. La distancia “correcta” siempre será la suma de las distancias cortas. Recuerda, El COD se calcula a base de la ley de porbabilidad de dos eventos independientes que ocurren al mismo tiempo.

Coeficiente de coincidencia e Interferencia Frecuencia observada 1 / 2, 205 = 0. 00045 Frecuencia esperada (frec. COS 1)(frec. COS 2) = (0. 0132)(0. 3260) = 0. 004 CC = Frec. Observada / Frec. Esperada = 0. 00045 / 0. 004 = 0. 225 x 100 = 22. 5% Los resultados indican que hubo interferencia, por lo tanto, los eventos no son independientes. ¿Cuánta interferencia hubo? Interferencia = 1 -CC = 1 -0. 225 = 0. 775 ó 77. 5% Recuerda, mientras más cercanos los genes, mayor la interferencia. La frecuencia de recombinación observada no excederá 50%. ¿Por qué?

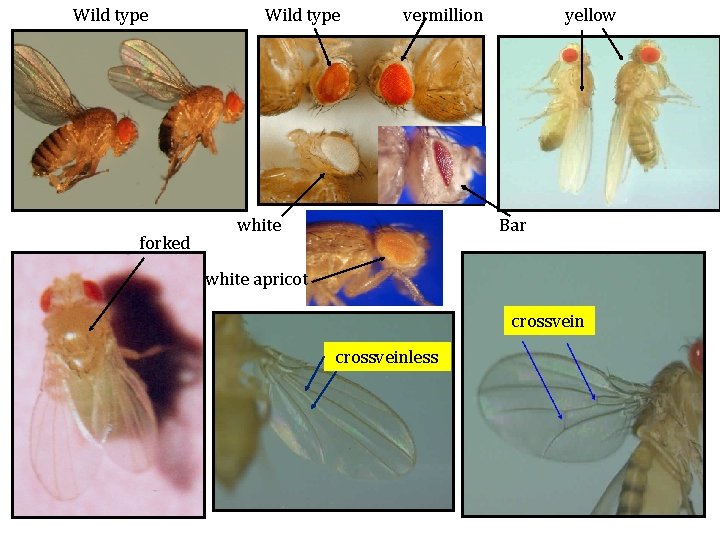
Wild type forked Wild type vermillion white yellow Bar white apricot crossveinless