Gene Structure and Identification Genes and Genomes ORFs







- Slides: 31

Gene Structure and Identification • • Genes and Genomes ORFs and more Consensus Sequences Gene Finding Reading: sections 1. 3, 9. 1 -9. 6 BIO 520 Bioinformatics Jim Lund

Gene The functional and physical unit of heredity passed from parent to offspring. Genes are pieces of DNA, and most genes contain the information for making a specific protein.

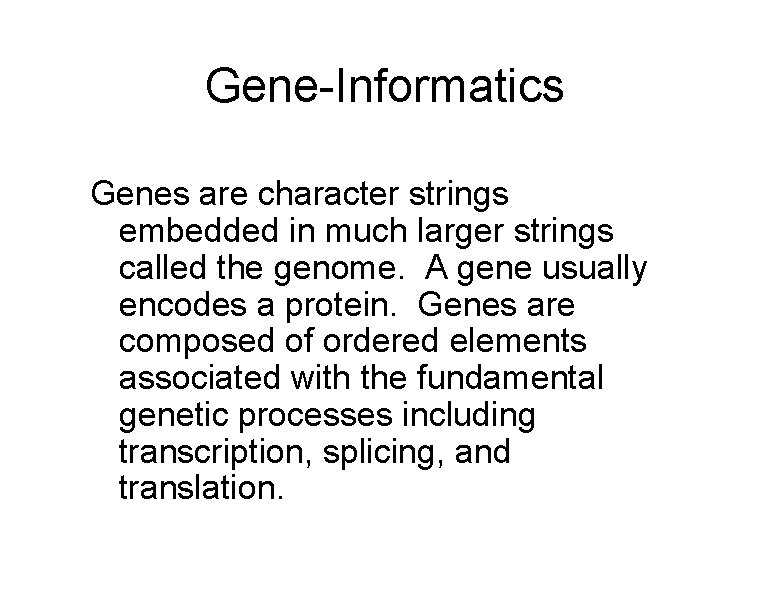
Gene-Informatics Genes are character strings embedded in much larger strings called the genome. A gene usually encodes a protein. Genes are composed of ordered elements associated with the fundamental genetic processes including transcription, splicing, and translation.

ACGT to Gene • Cells recognize genes from DNA sequence.

Genes • Protein Coding • RNA genes – r. RNA – t. RNA – si. RNA, mi. RNA, sno. RNA…

Genomes • Genome seq. has only limited use by itself – Markers, SNPs, etc. • Functional annotation – Identify proteins and their functions. – And regulatory regions, etc. • Parts list: a source for understanding all biology--and ushers in the post-genomic age of biology.

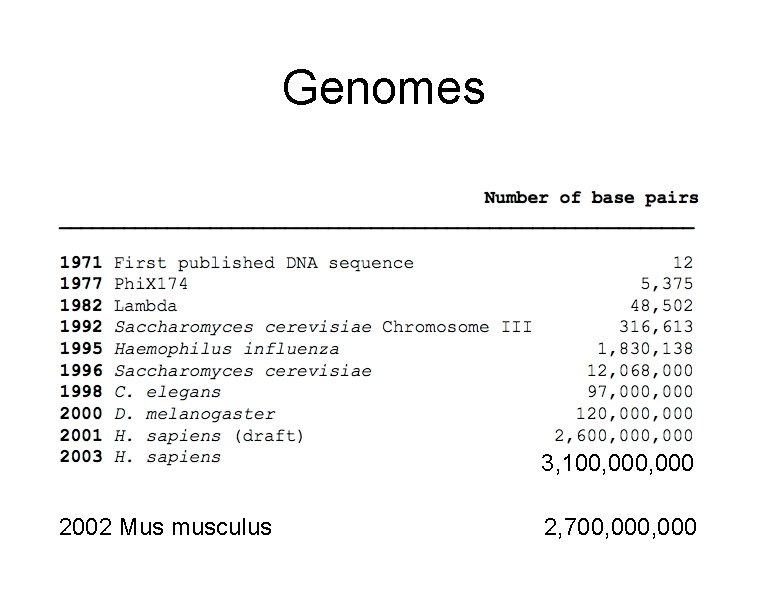
Genomes 3, 100, 000 2002 Mus musculus 2, 700, 000

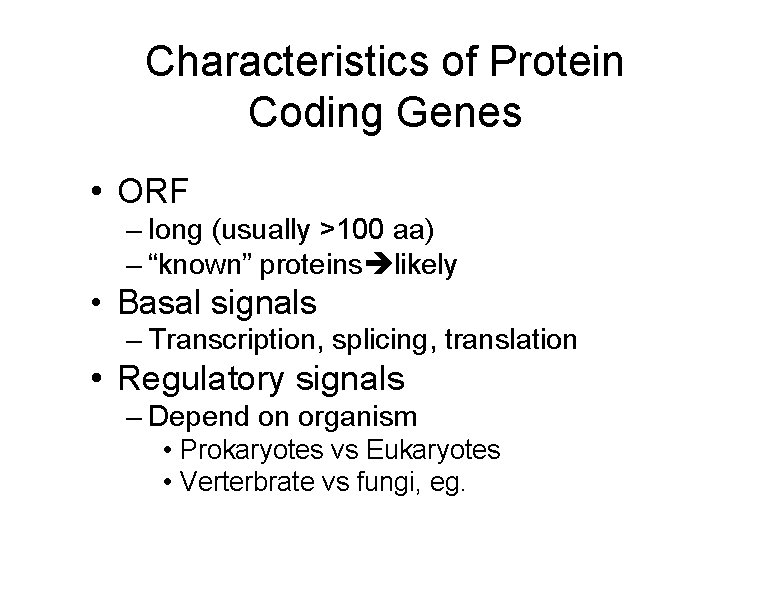
Characteristics of Protein Coding Genes • ORF – long (usually >100 aa) – “known” proteins likely • Basal signals – Transcription, splicing, translation • Regulatory signals – Depend on organism • Prokaryotes vs Eukaryotes • Verterbrate vs fungi, eg.

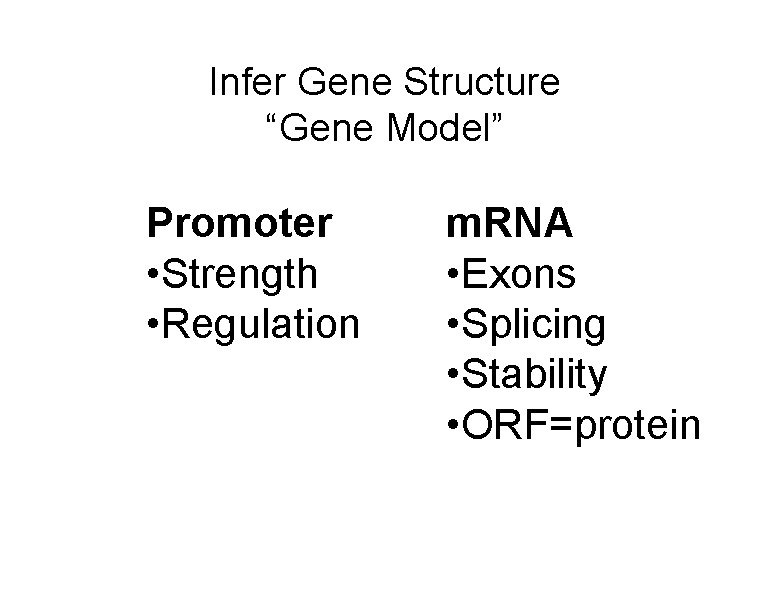
Infer Gene Structure “Gene Model” Promoter • Strength • Regulation m. RNA • Exons • Splicing • Stability • ORF=protein

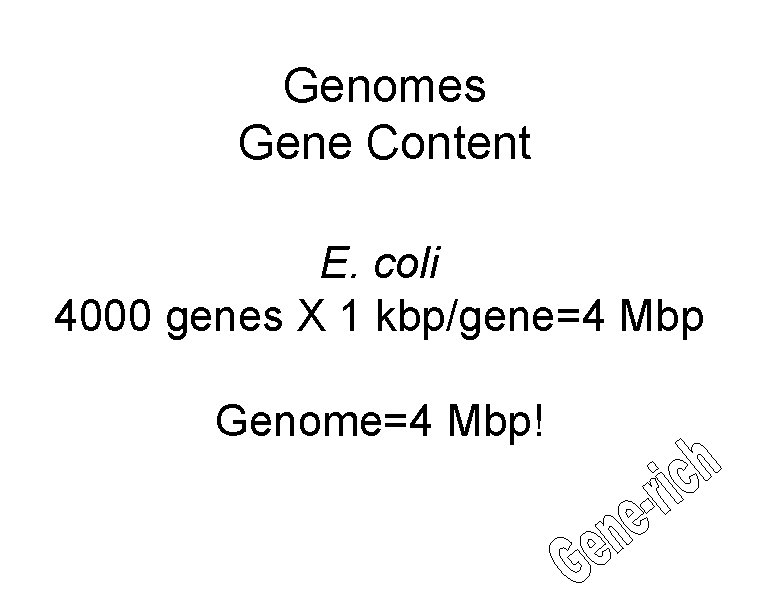
Genomes Gene Content E. coli 4000 genes X 1 kbp/gene=4 Mbp Genome=4 Mbp!

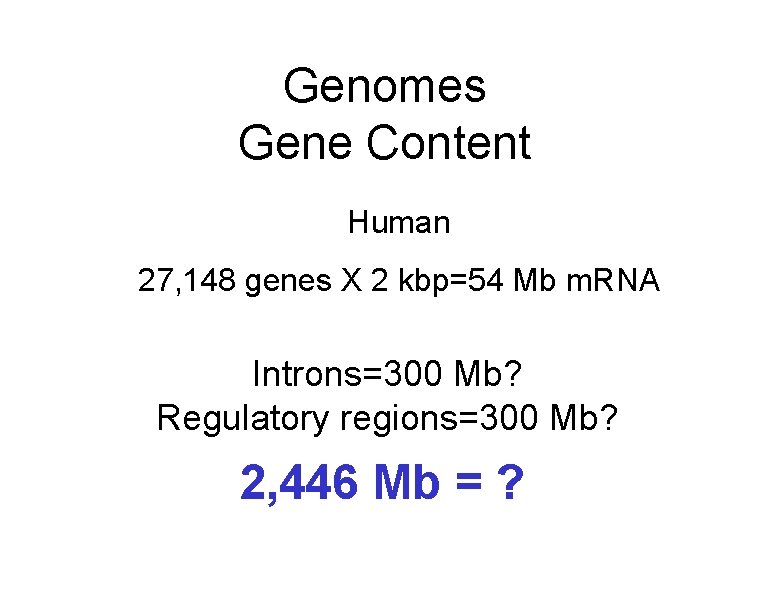
Genomes Gene Content Human 27, 148 genes X 2 kbp=54 Mb m. RNA Introns=300 Mb? Regulatory regions=300 Mb? 2, 446 Mb = ?

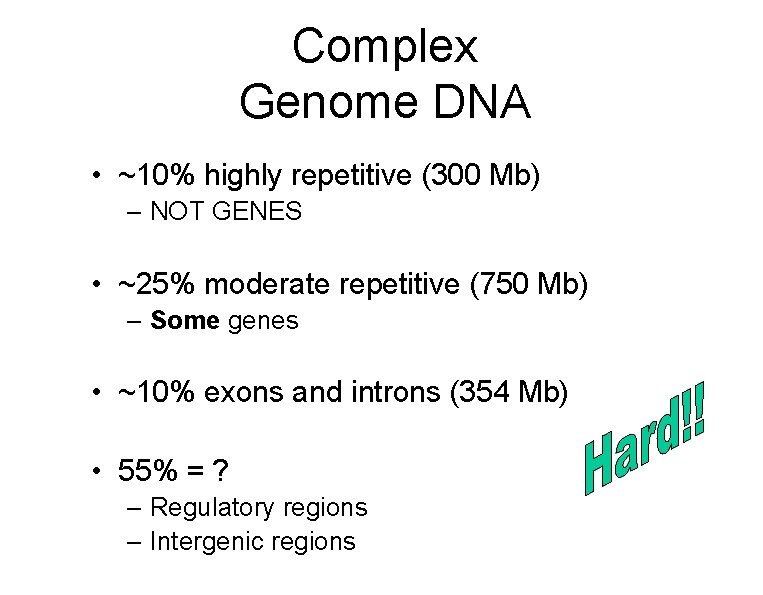
Complex Genome DNA • ~10% highly repetitive (300 Mb) – NOT GENES • ~25% moderate repetitive (750 Mb) – Some genes • ~10% exons and introns (354 Mb) • 55% = ? – Regulatory regions – Intergenic regions

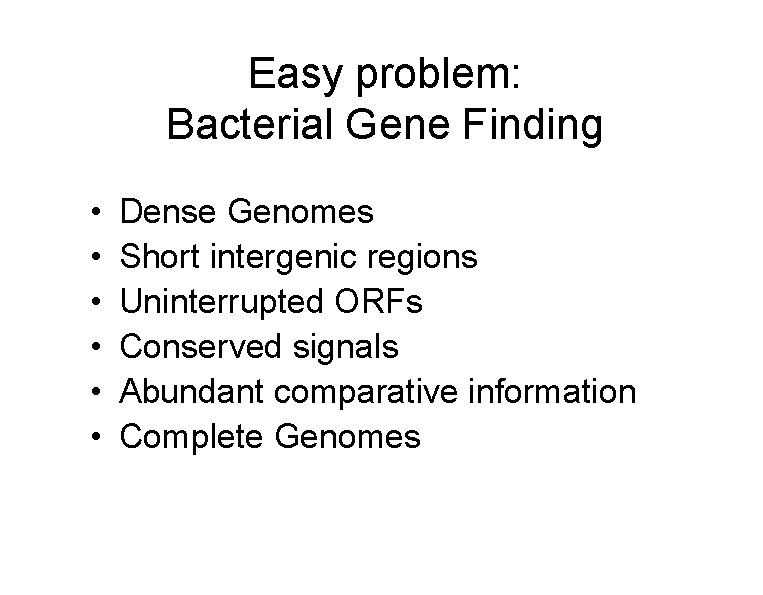
Easy problem: Bacterial Gene Finding • • • Dense Genomes Short intergenic regions Uninterrupted ORFs Conserved signals Abundant comparative information Complete Genomes

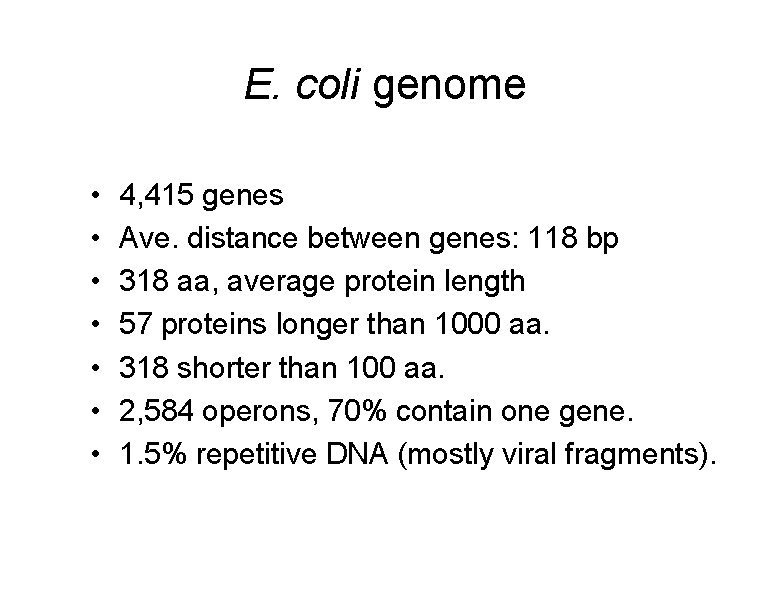
E. coli genome • • 4, 415 genes Ave. distance between genes: 118 bp 318 aa, average protein length 57 proteins longer than 1000 aa. 318 shorter than 100 aa. 2, 584 operons, 70% contain one gene. 1. 5% repetitive DNA (mostly viral fragments).

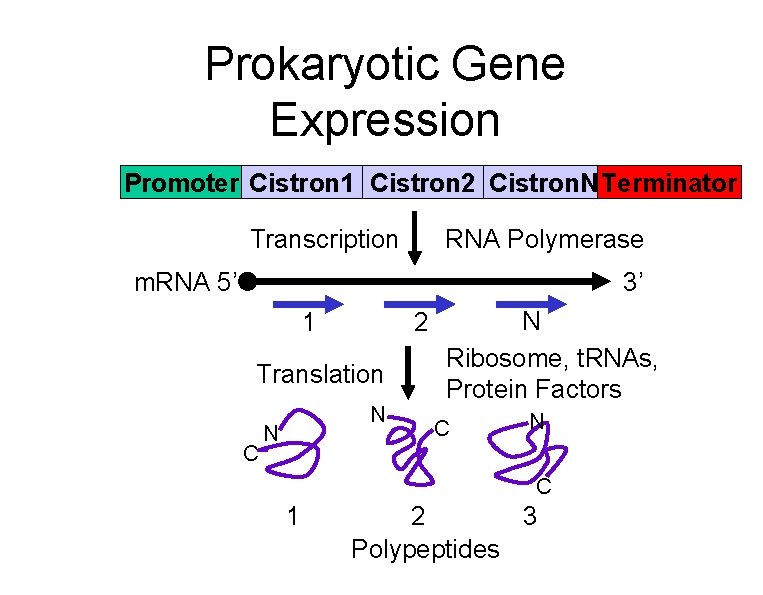
Prokaryotic Gene Expression Promoter Cistron 1 Cistron 2 Cistron. N Terminator Transcription RNA Polymerase m. RNA 5’ 3’ 1 2 Translation C N N N Ribosome, t. RNAs, Protein Factors C N C 1 2 3 Polypeptides

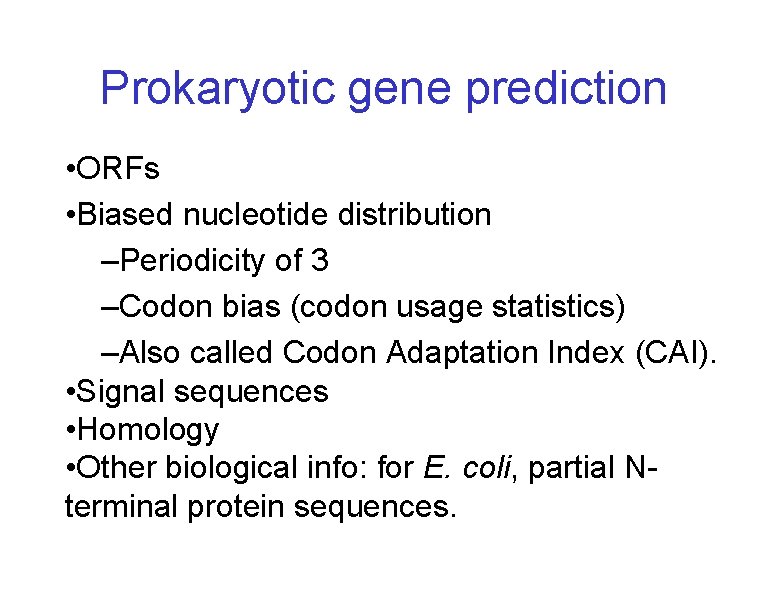
Prokaryotic gene prediction • ORFs • Biased nucleotide distribution –Periodicity of 3 –Codon bias (codon usage statistics) –Also called Codon Adaptation Index (CAI). • Signal sequences • Homology • Other biological info: for E. coli, partial Nterminal protein sequences.

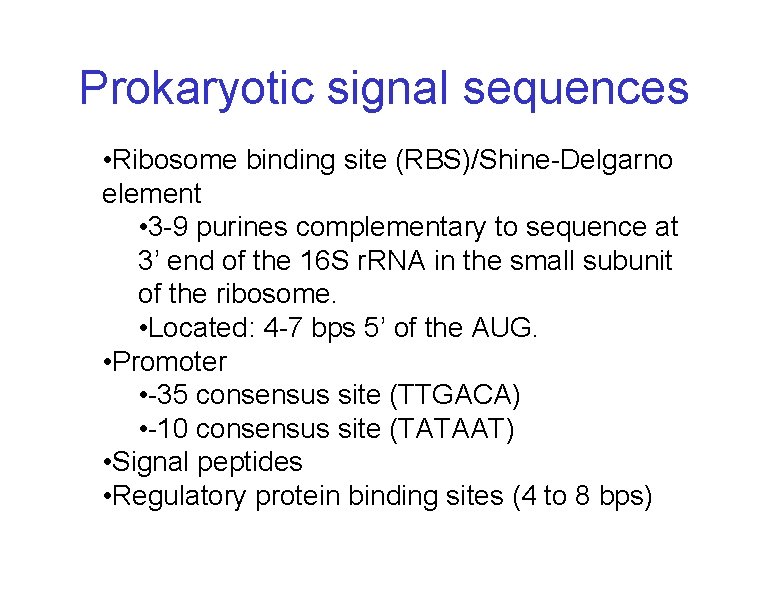
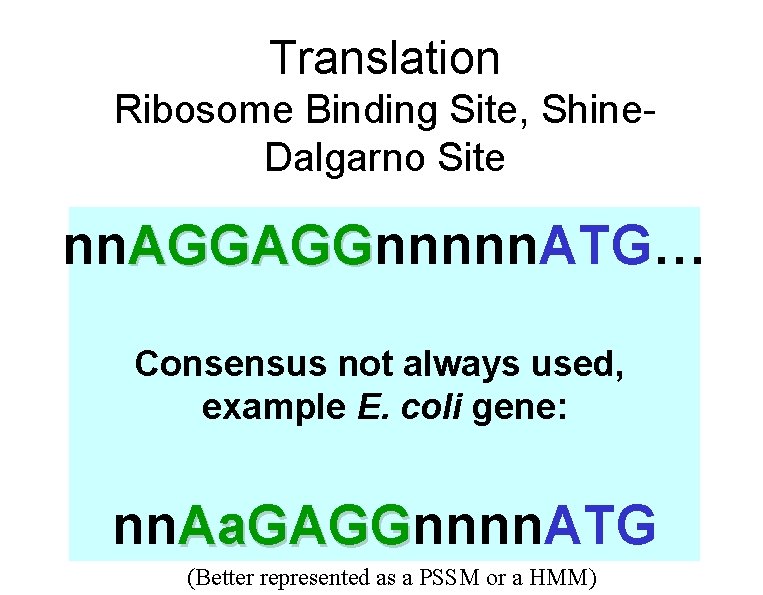
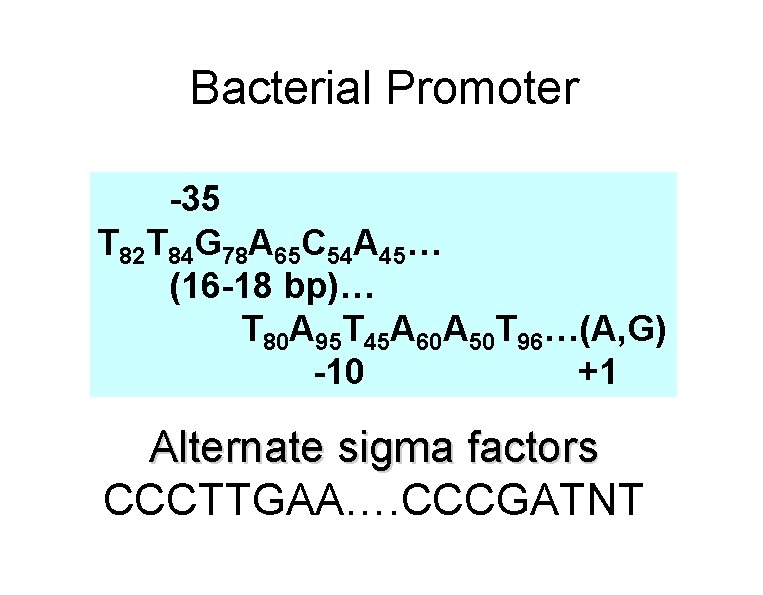
Prokaryotic signal sequences • Ribosome binding site (RBS)/Shine-Delgarno element • 3 -9 purines complementary to sequence at 3’ end of the 16 S r. RNA in the small subunit of the ribosome. • Located: 4 -7 bps 5’ of the AUG. • Promoter • -35 consensus site (TTGACA) • -10 consensus site (TATAAT) • Signal peptides • Regulatory protein binding sites (4 to 8 bps)

ORFs n P(ORF)=(61/64) 20 P(20)=(61/64) =. 38 P(100)=0. 008 -4 P(200)=10

ORF finding tools • Artemis – analyze ORFs • • Testcode (Fickett’s) Codon. Preference ORF Finder (NCBI) BCM Search Launcher

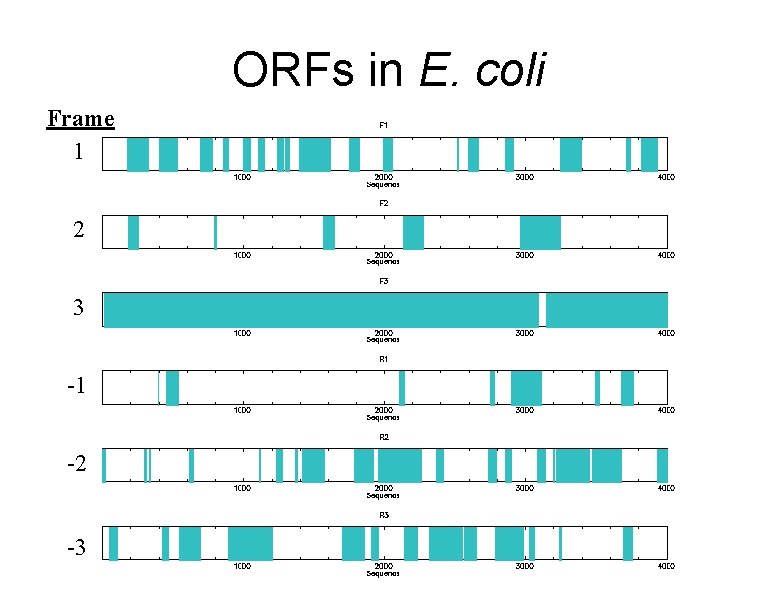
ORFs in E. coli Frame 1 2 3 -1 -2 -3

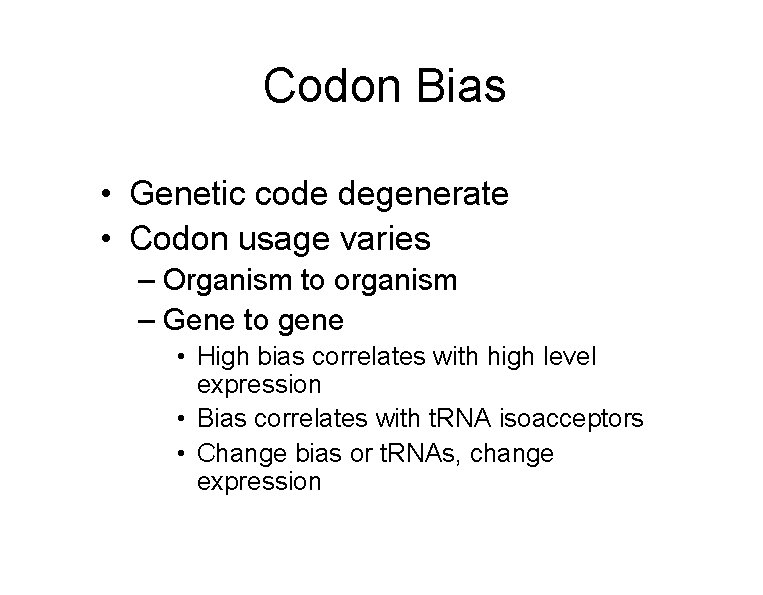
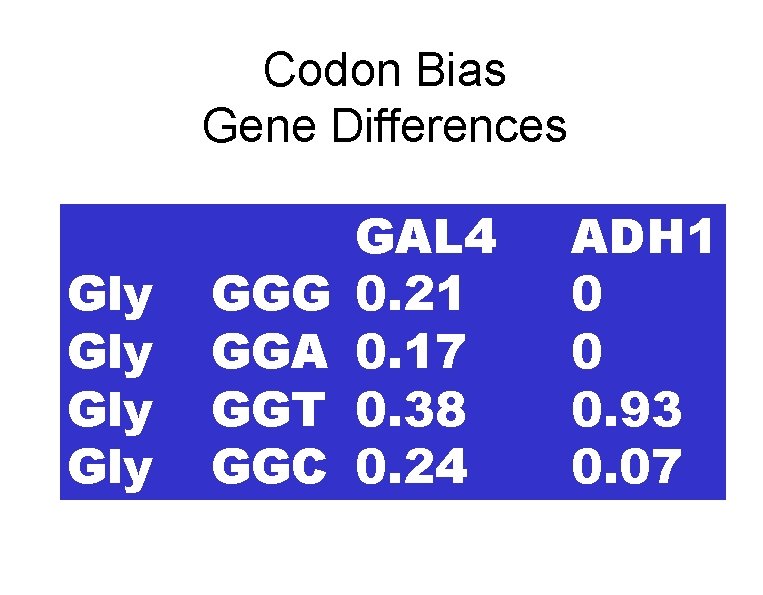
Codon Bias • Genetic code degenerate • Codon usage varies – Organism to organism – Gene to gene • High bias correlates with high level expression • Bias correlates with t. RNA isoacceptors • Change bias or t. RNAs, change expression

Codon Bias Gly Gly GGG GGA GGT GGC 6 6 0. 21 0. 17 0. 38 0. 24

Codon Bias Gene Differences Gly Gly GGG GGA GGT GGC GAL 4 0. 21 0. 17 0. 38 0. 24 ADH 1 0 0 0. 93 0. 07

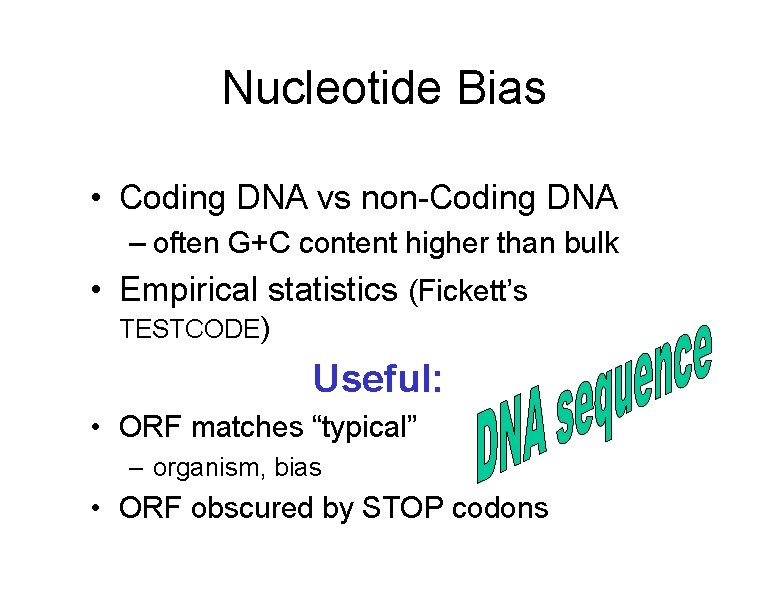
Nucleotide Bias • Coding DNA vs non-Coding DNA – often G+C content higher than bulk • Empirical statistics (Fickett’s TESTCODE) Useful: • ORF matches “typical” – organism, bias • ORF obscured by STOP codons

We found ORFs-now what? • Work backwards – Locate adjacent cistrons – Locate RBS – Locate promoter – Locate terminator – Locate regulatory sites

Operon Structure Promoter?

Translation Ribosome Binding Site, Shine. Dalgarno Site nn. AGGAGGnnnnn. ATG… AGGAGG Consensus not always used, example E. coli gene: nn. Aa. GAGGnnnn. ATG Aa. GAGG (Better represented as a PSSM or a HMM)

Bacterial Promoter -35 T 82 T 84 G 78 A 65 C 54 A 45… (16 -18 bp)… T 80 A 95 T 45 A 60 A 50 T 96…(A, G) -10 +1 Alternate sigma factors CCCTTGAA…. CCCGATNT

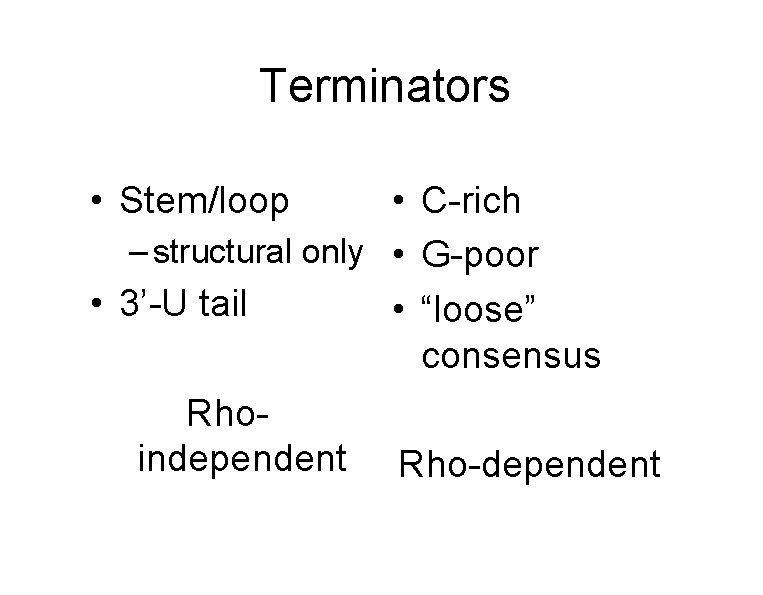
Terminators • Stem/loop • C-rich – structural only • G-poor • 3’-U tail • “loose” consensus Rhoindependent Rho-dependent

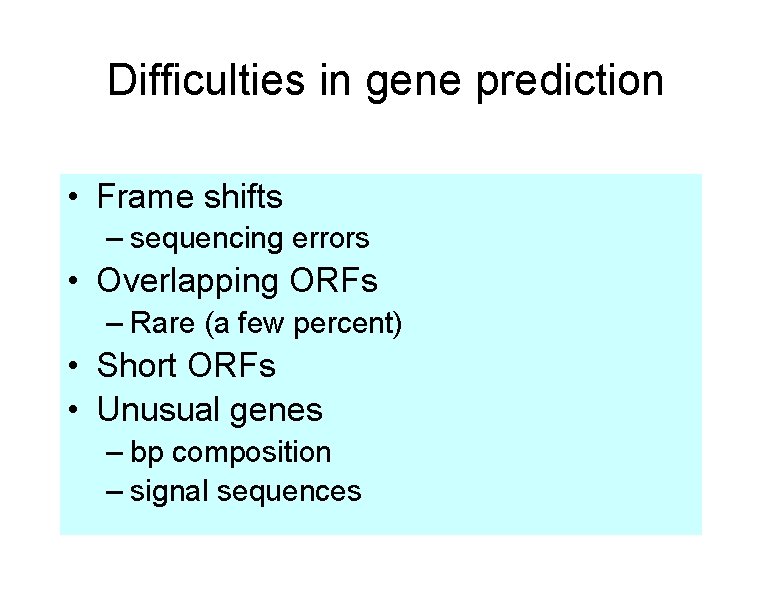
Difficulties in gene prediction • Frame shifts – sequencing errors • Overlapping ORFs – Rare (a few percent) • Short ORFs • Unusual genes – bp composition – signal sequences

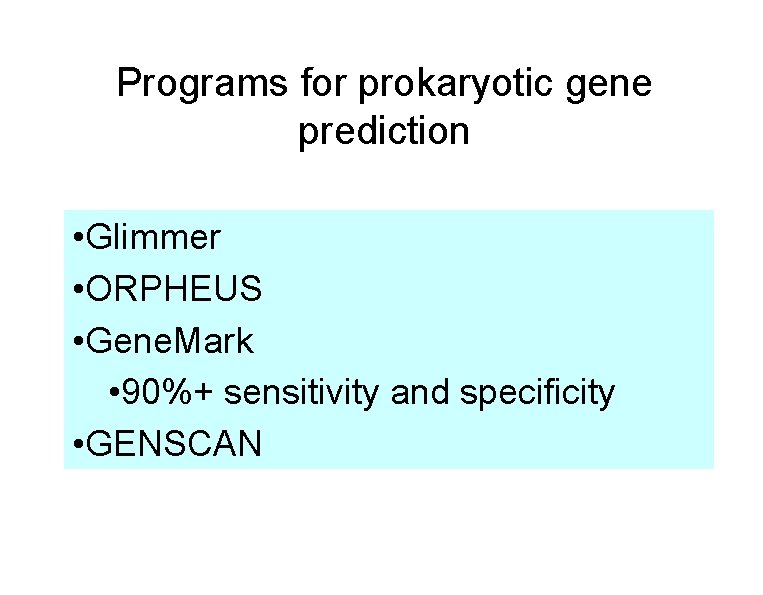
Programs for prokaryotic gene prediction • Glimmer • ORPHEUS • Gene. Mark • 90%+ sensitivity and specificity • GENSCAN