Fig 1 Splice site composition in Ectocarpus Genome
- Slides: 10

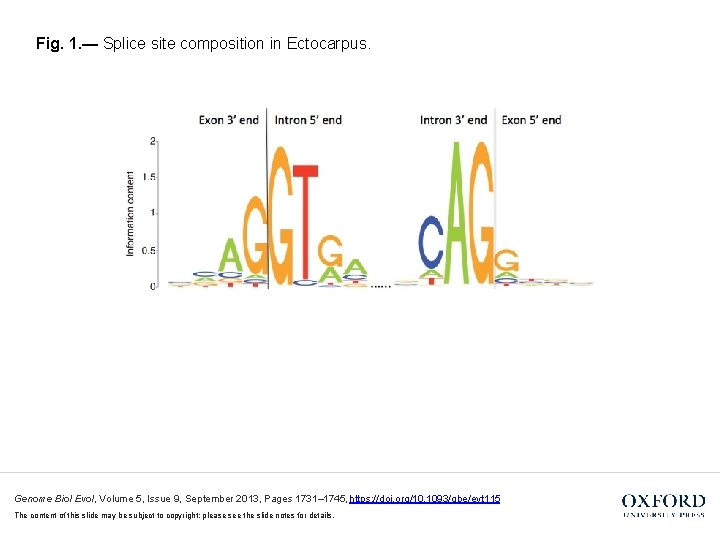
Fig. 1. — Splice site composition in Ectocarpus. Genome Biol Evol, Volume 5, Issue 9, September 2013, Pages 1731– 1745, https: //doi. org/10. 1093/gbe/evt 115 The content of this slide may be subject to copyright: please see the slide notes for details.

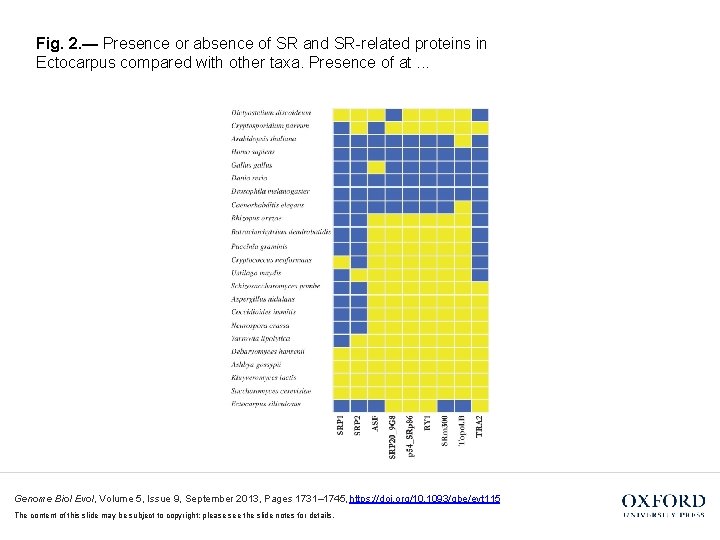
Fig. 2. — Presence or absence of SR and SR-related proteins in Ectocarpus compared with other taxa. Presence of at. . . Genome Biol Evol, Volume 5, Issue 9, September 2013, Pages 1731– 1745, https: //doi. org/10. 1093/gbe/evt 115 The content of this slide may be subject to copyright: please see the slide notes for details.

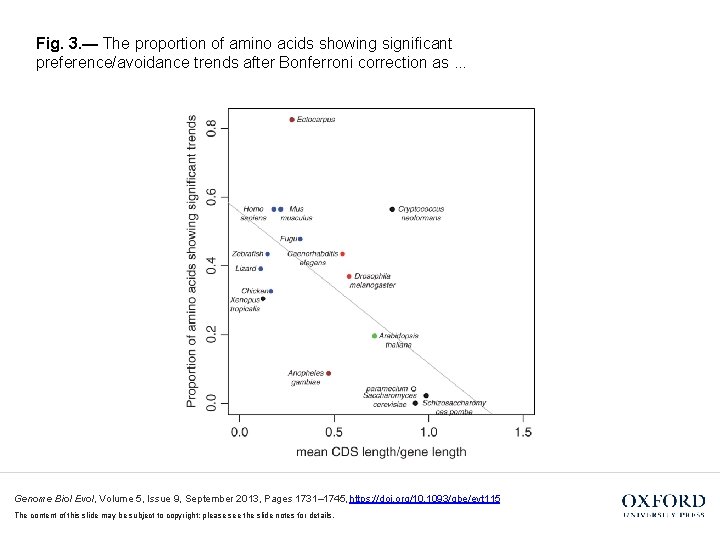
Fig. 3. — The proportion of amino acids showing significant preference/avoidance trends after Bonferroni correction as. . . Genome Biol Evol, Volume 5, Issue 9, September 2013, Pages 1731– 1745, https: //doi. org/10. 1093/gbe/evt 115 The content of this slide may be subject to copyright: please see the slide notes for details.

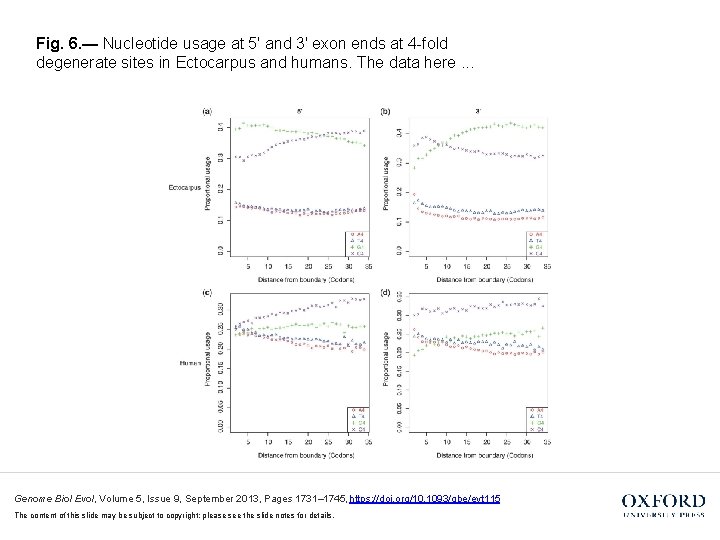
Fig. 6. — Nucleotide usage at 5′ and 3′ exon ends at 4 -fold degenerate sites in Ectocarpus and humans. The data here. . . Genome Biol Evol, Volume 5, Issue 9, September 2013, Pages 1731– 1745, https: //doi. org/10. 1093/gbe/evt 115 The content of this slide may be subject to copyright: please see the slide notes for details.

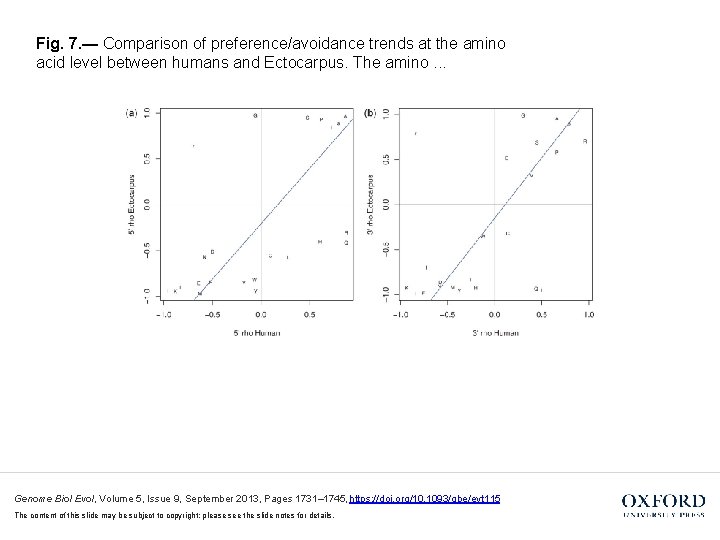
Fig. 7. — Comparison of preference/avoidance trends at the amino acid level between humans and Ectocarpus. The amino. . . Genome Biol Evol, Volume 5, Issue 9, September 2013, Pages 1731– 1745, https: //doi. org/10. 1093/gbe/evt 115 The content of this slide may be subject to copyright: please see the slide notes for details.

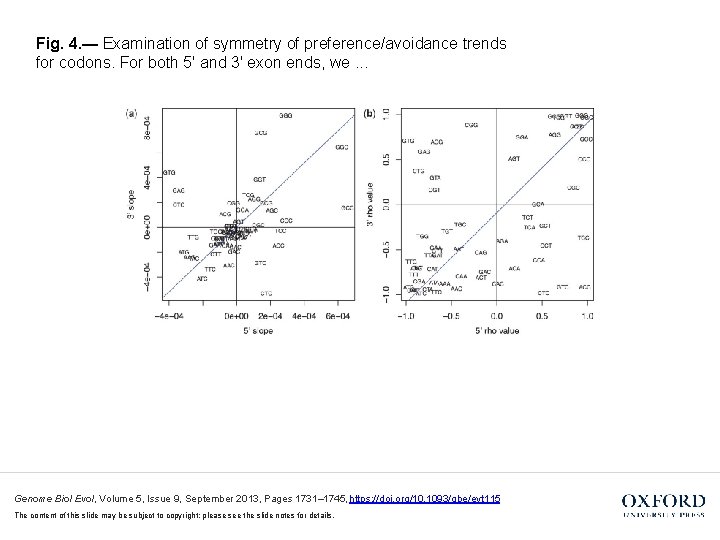
Fig. 4. — Examination of symmetry of preference/avoidance trends for codons. For both 5′ and 3′ exon ends, we. . . Genome Biol Evol, Volume 5, Issue 9, September 2013, Pages 1731– 1745, https: //doi. org/10. 1093/gbe/evt 115 The content of this slide may be subject to copyright: please see the slide notes for details.

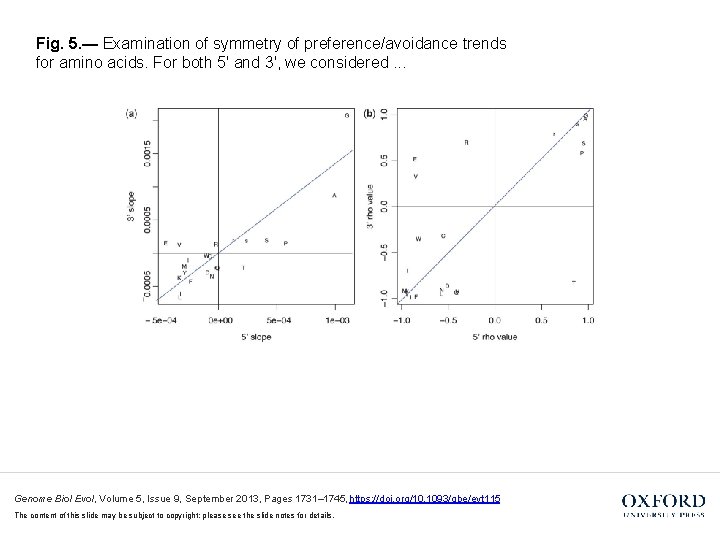
Fig. 5. — Examination of symmetry of preference/avoidance trends for amino acids. For both 5′ and 3′, we considered. . . Genome Biol Evol, Volume 5, Issue 9, September 2013, Pages 1731– 1745, https: //doi. org/10. 1093/gbe/evt 115 The content of this slide may be subject to copyright: please see the slide notes for details.

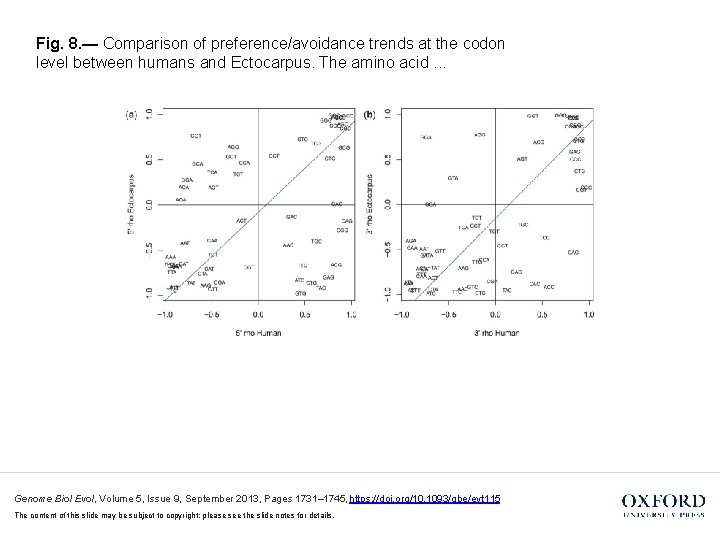
Fig. 8. — Comparison of preference/avoidance trends at the codon level between humans and Ectocarpus. The amino acid. . . Genome Biol Evol, Volume 5, Issue 9, September 2013, Pages 1731– 1745, https: //doi. org/10. 1093/gbe/evt 115 The content of this slide may be subject to copyright: please see the slide notes for details.

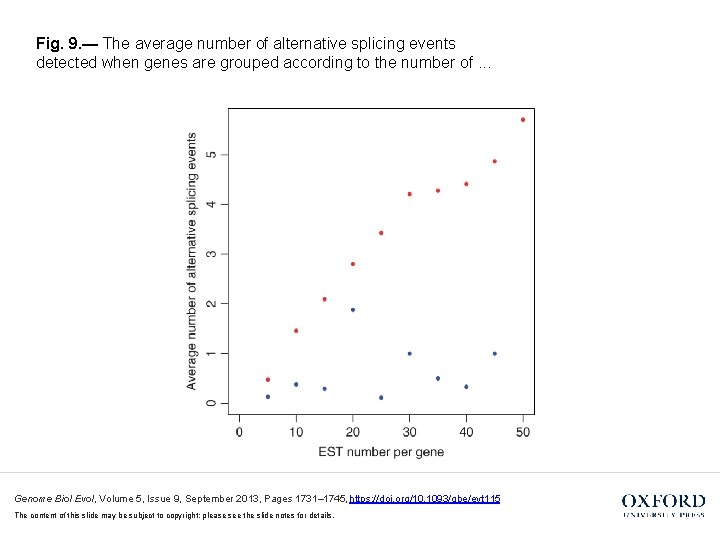
Fig. 9. — The average number of alternative splicing events detected when genes are grouped according to the number of. . . Genome Biol Evol, Volume 5, Issue 9, September 2013, Pages 1731– 1745, https: //doi. org/10. 1093/gbe/evt 115 The content of this slide may be subject to copyright: please see the slide notes for details.

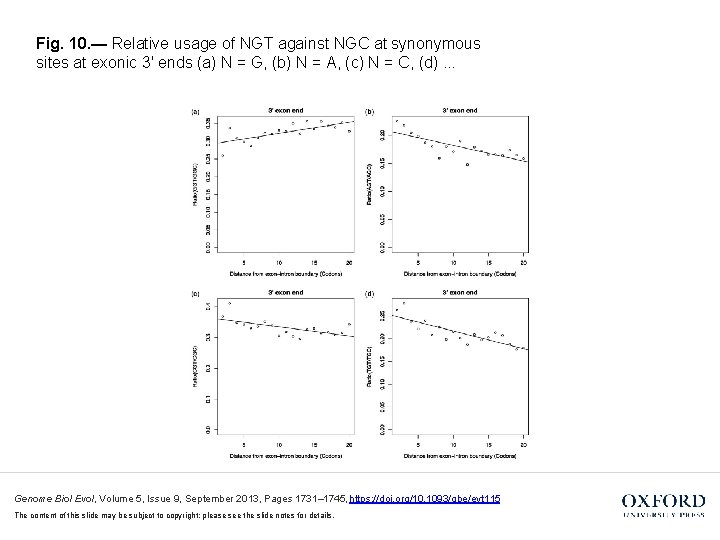
Fig. 10. — Relative usage of NGT against NGC at synonymous sites at exonic 3′ ends (a) N = G, (b) N = A, (c) N = C, (d). . . Genome Biol Evol, Volume 5, Issue 9, September 2013, Pages 1731– 1745, https: //doi. org/10. 1093/gbe/evt 115 The content of this slide may be subject to copyright: please see the slide notes for details.