DNA Structure Replication DNA Structure Chromosomes are made



- Slides: 34

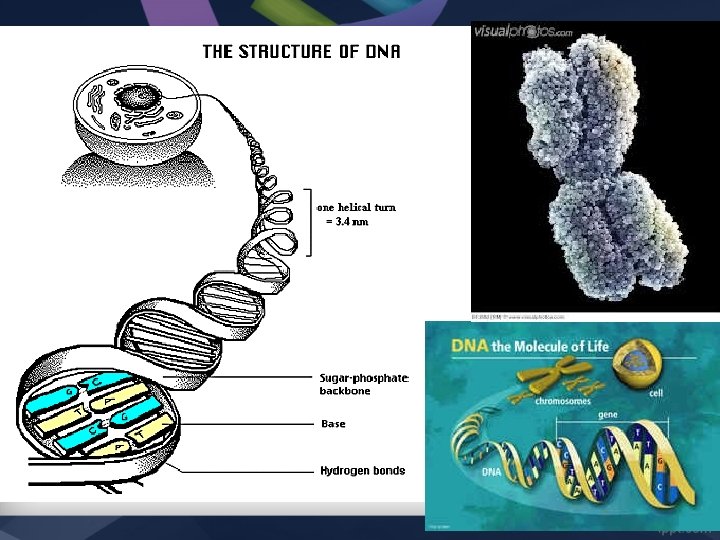
DNA Structure & Replication


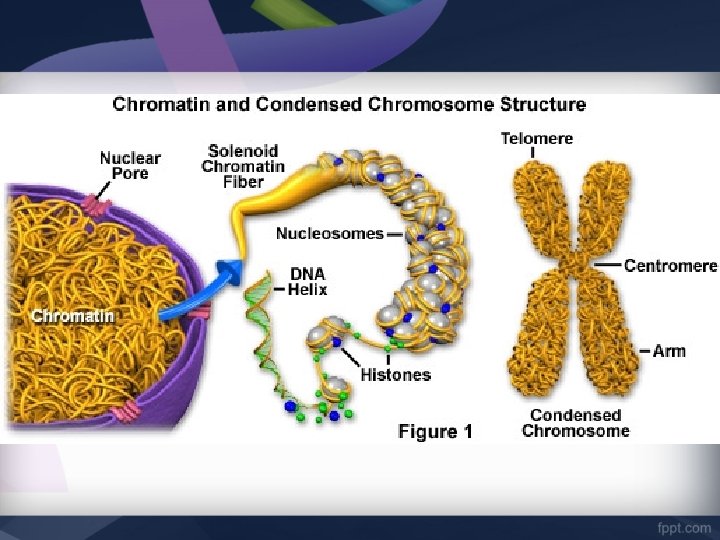
DNA Structure • Chromosomes are made of DNA. • The DNA is wound around proteins called Histones • When chromosomes are unraveled, the DNA strands are called Chromatin • DNA is made of Nucleotides – Deoxyribose (5 carbon sugar) – Phosphate group – Nitrogen Base (A, T, G, C)


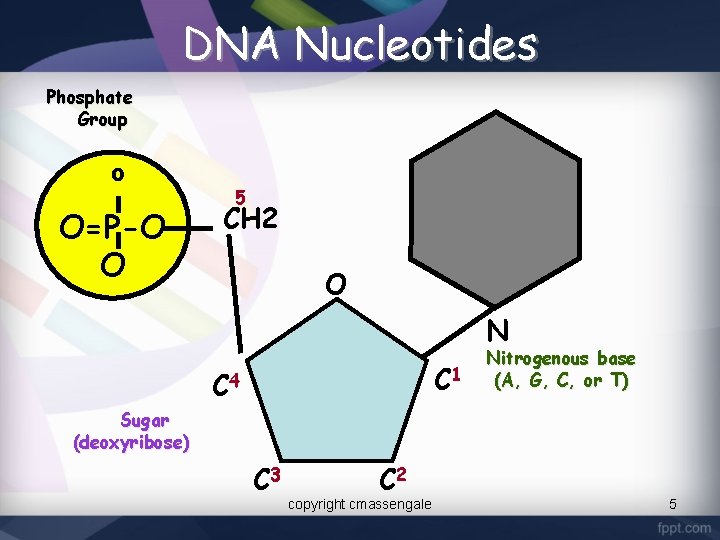
DNA Nucleotides Phosphate Group O O=P-O O 5 CH 2 O N C 1 C 4 Nitrogenous base (A, G, C, or T) Sugar (deoxyribose) C 3 C 2 copyright cmassengale 5

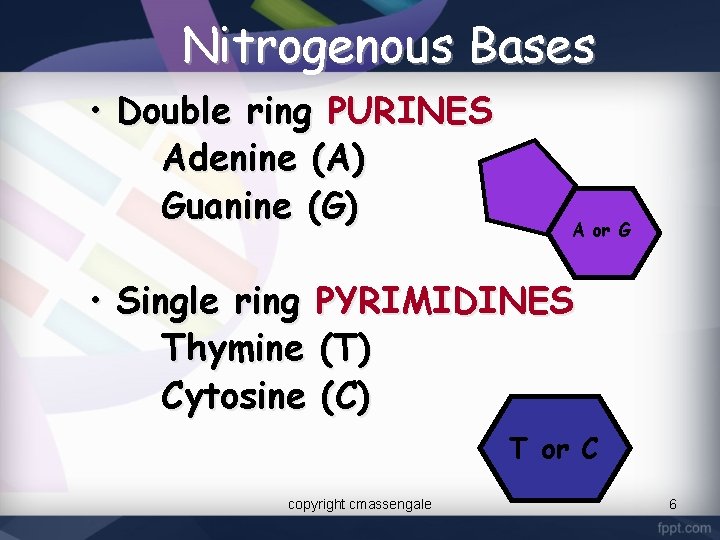
Nitrogenous Bases • Double ring PURINES Adenine (A) Guanine (G) A or G • Single ring PYRIMIDINES Thymine (T) Cytosine (C) T or C copyright cmassengale 6

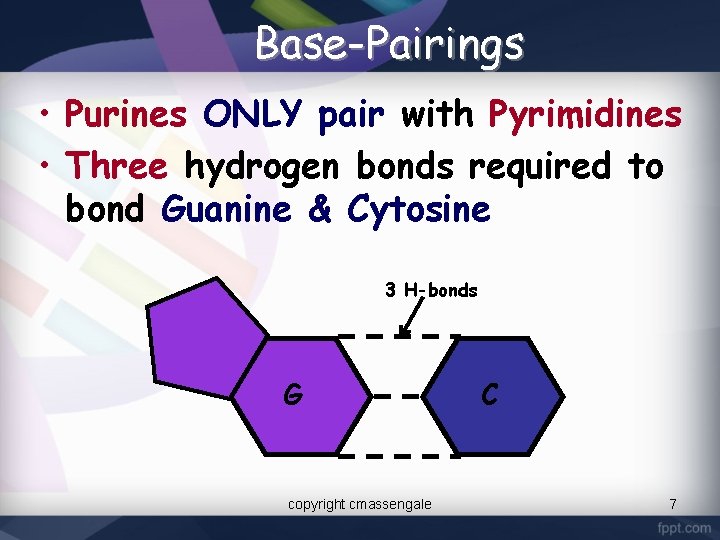
Base-Pairings • Purines ONLY pair with Pyrimidines • Three hydrogen bonds required to bond Guanine & Cytosine 3 H-bonds G copyright cmassengale C 7

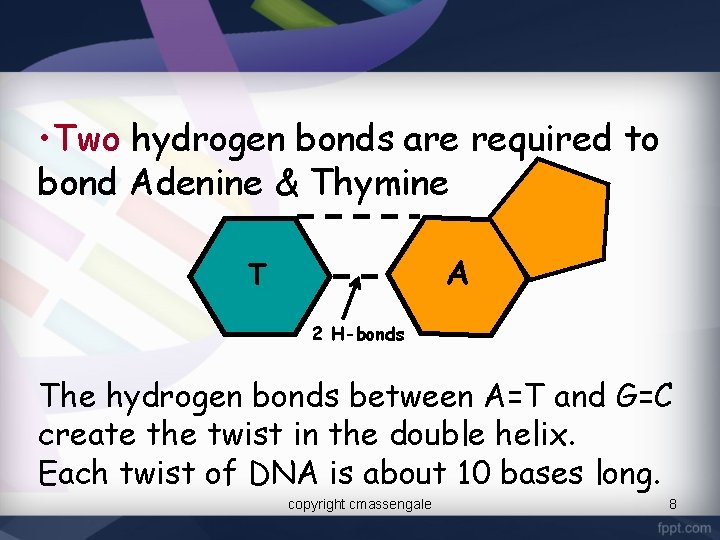
• Two hydrogen bonds are required to bond Adenine & Thymine A T 2 H-bonds The hydrogen bonds between A=T and G=C create the twist in the double helix. Each twist of DNA is about 10 bases long. copyright cmassengale 8

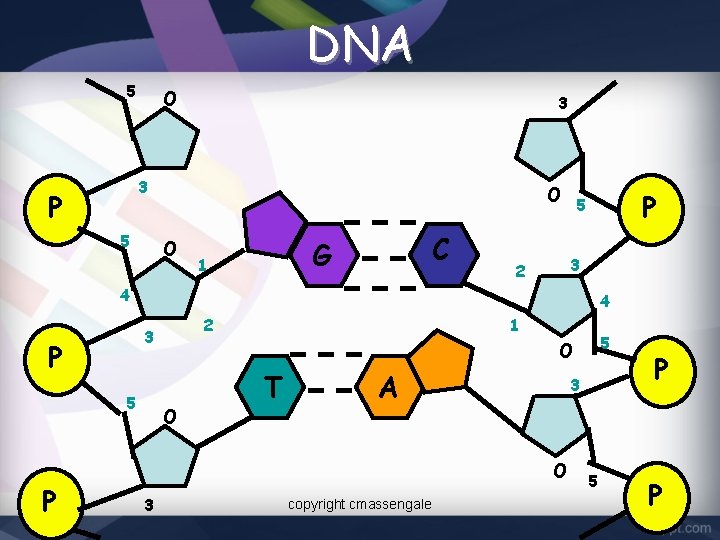
DNA 5 O 3 3 P 5 O O C G 1 P 5 3 2 4 4 P 5 P 2 3 1 O T A 3 O 3 5 O copyright cmassengale 5 P P 9

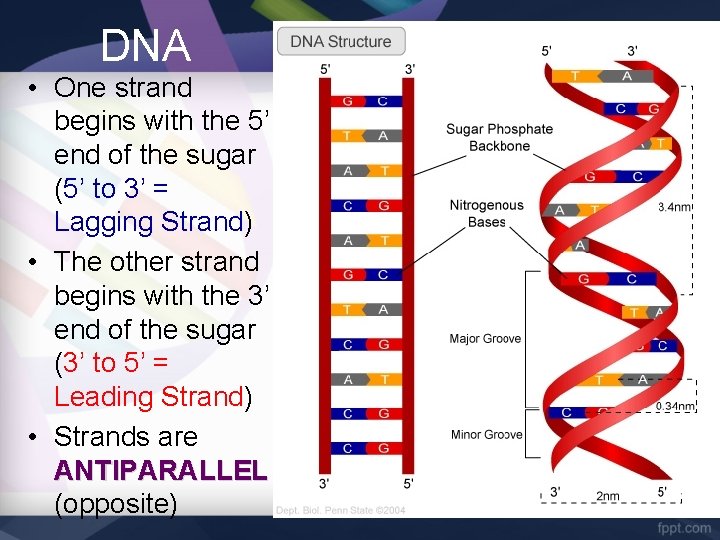
DNA • One strand begins with the 5’ end of the sugar (5’ to 3’ = Lagging Strand) • The other strand begins with the 3’ end of the sugar (3’ to 5’ = Leading Strand) • Strands are ANTIPARALLEL (opposite)

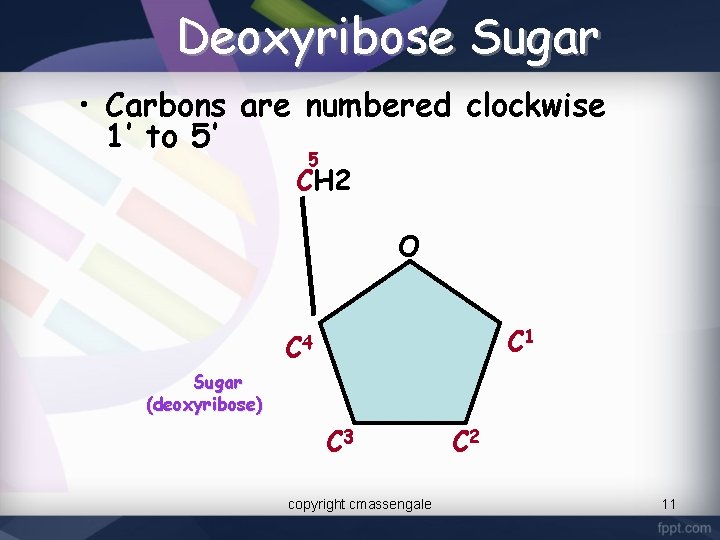
Deoxyribose Sugar • Carbons are numbered clockwise 1’ to 5’ 5 CH 2 O C 1 C 4 Sugar (deoxyribose) C 3 copyright cmassengale C 2 11

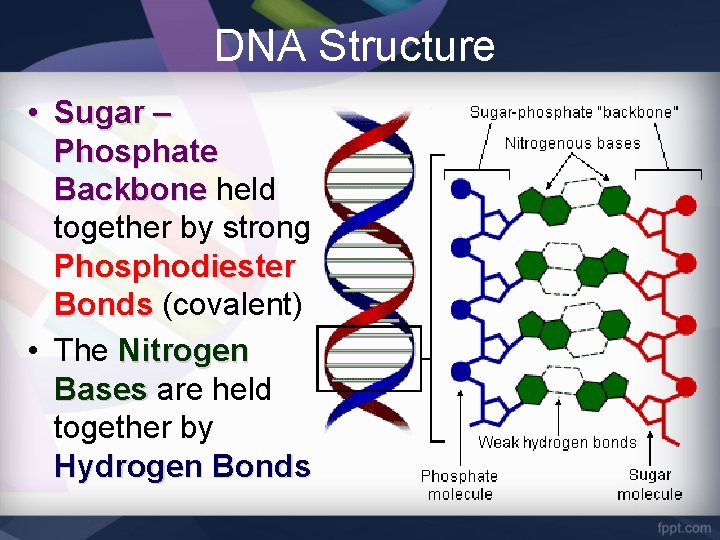
DNA Structure • Sugar – Phosphate Backbone held together by strong Phosphodiester Bonds (covalent) • The Nitrogen Bases are held together by Hydrogen Bonds

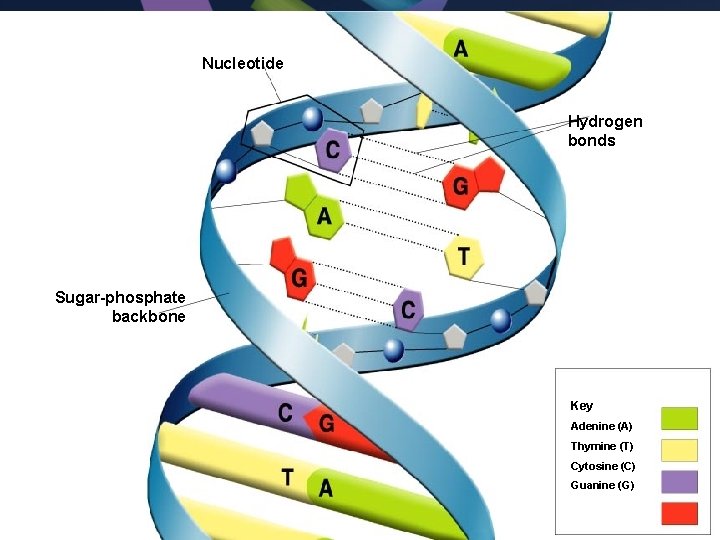
Nucleotide Hydrogen bonds Sugar-phosphate backbone Key Adenine (A) Thymine (T) Cytosine (C) Guanine (G)

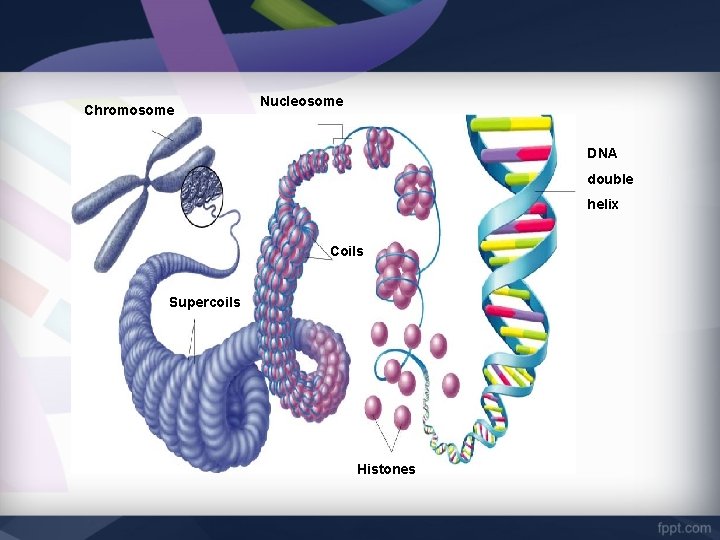
Chromosome Nucleosome DNA double helix Coils Supercoils Histones

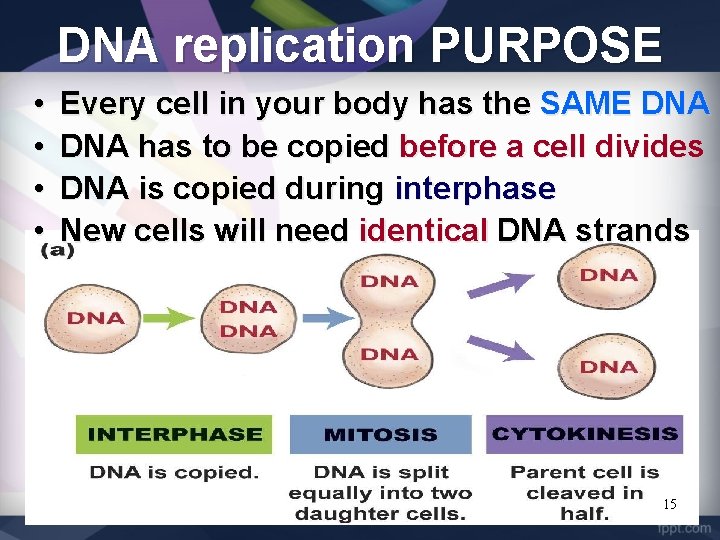
DNA replication PURPOSE • • Every cell in your body has the SAME DNA has to be copied before a cell divides DNA is copied during interphase New cells will need identical DNA strands 15

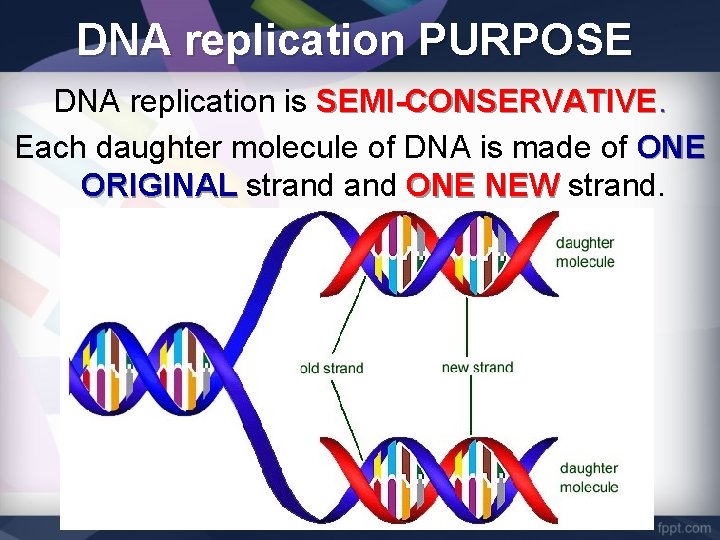
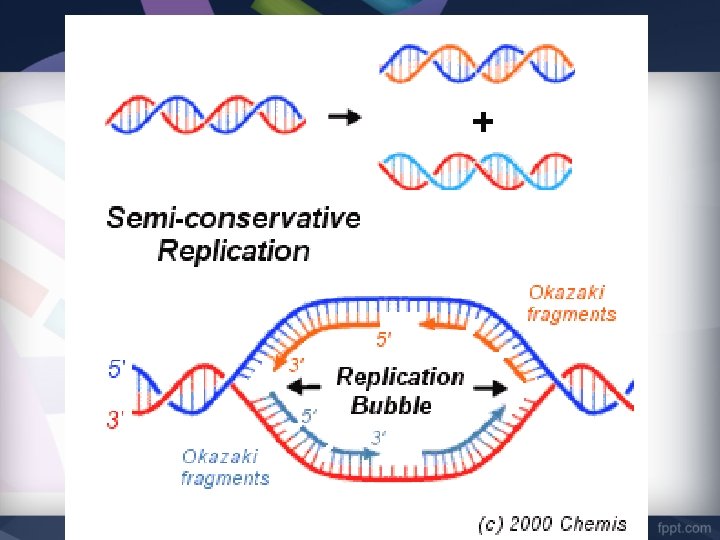
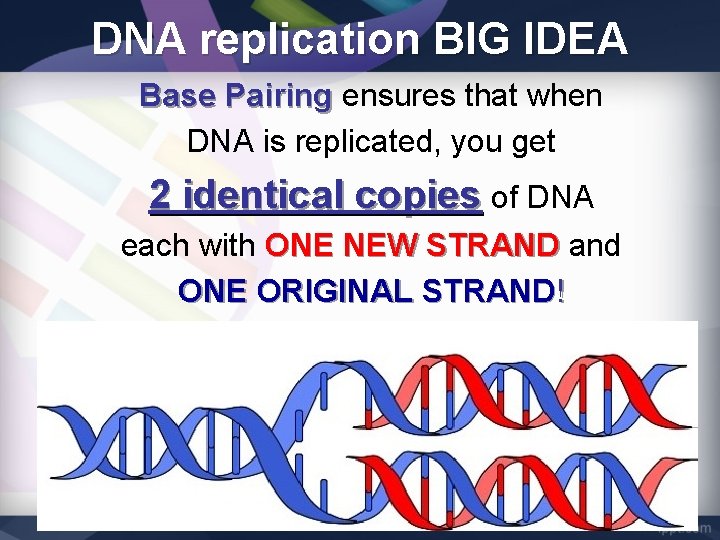
DNA replication PURPOSE DNA replication is SEMI-CONSERVATIVE. Each daughter molecule of DNA is made of ONE ORIGINAL strand ONE NEW strand.


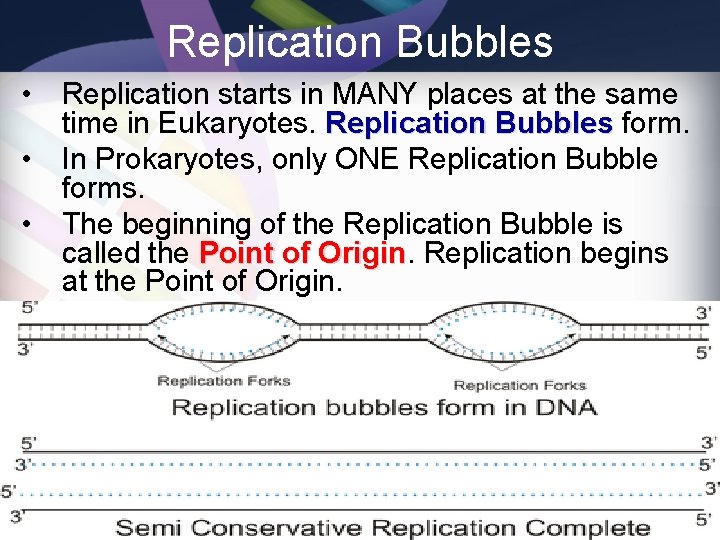
Replication Bubbles • Replication starts in MANY places at the same time in Eukaryotes. Replication Bubbles form. • In Prokaryotes, only ONE Replication Bubble forms. • The beginning of the Replication Bubble is called the Point of Origin Replication begins at the Point of Origin.

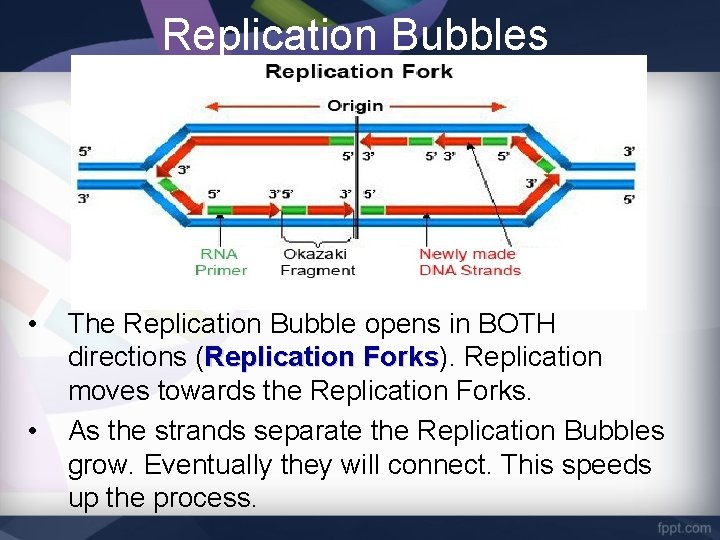
Replication Bubbles • • The Replication Bubble opens in BOTH directions (Replication Forks). Forks Replication moves towards the Replication Forks. As the strands separate the Replication Bubbles grow. Eventually they will connect. This speeds up the process.

Bell Ringer • If there is 30% Adenine in a human genome (DNA), how much Cytosine is present? Explain or Show your work.

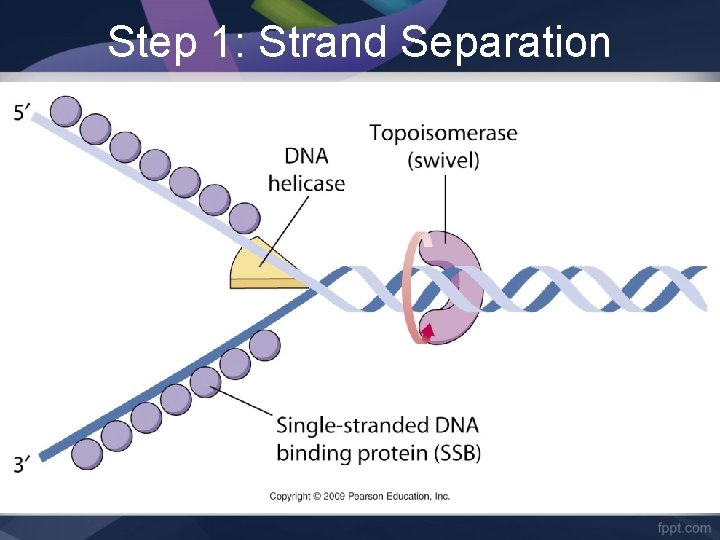
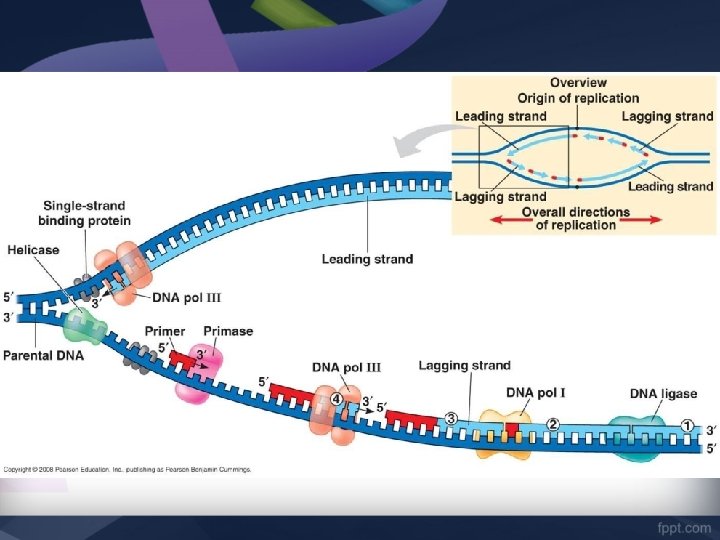
Step 1: Strand Separation 1. Helicase unwinds and separates the 2 DNA strands. 2. Single-Strand Binding proteins attach to the DNA strands to keep the strands from twisting. 3. Topoisomerase (another protein) attaches to the strands to relieve stress and keep the strands from breaking during replication.

Step 1: Strand Separation

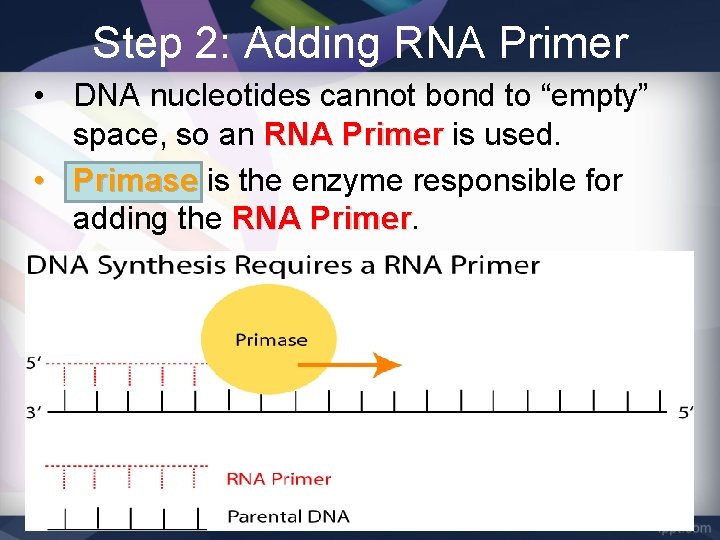
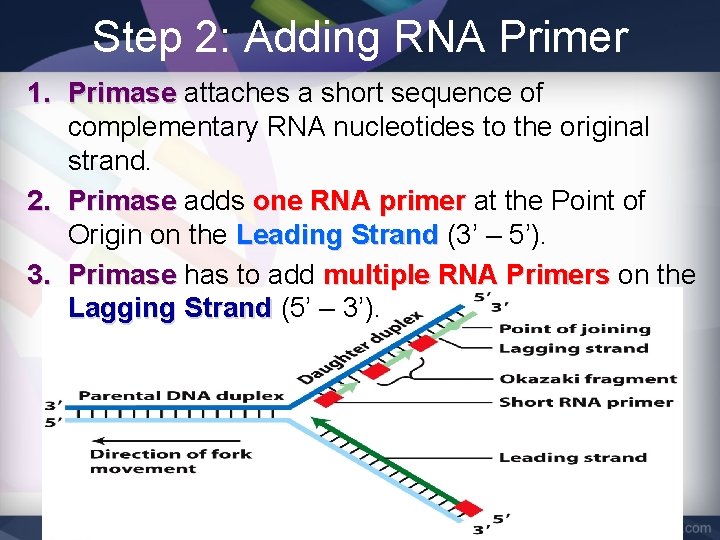
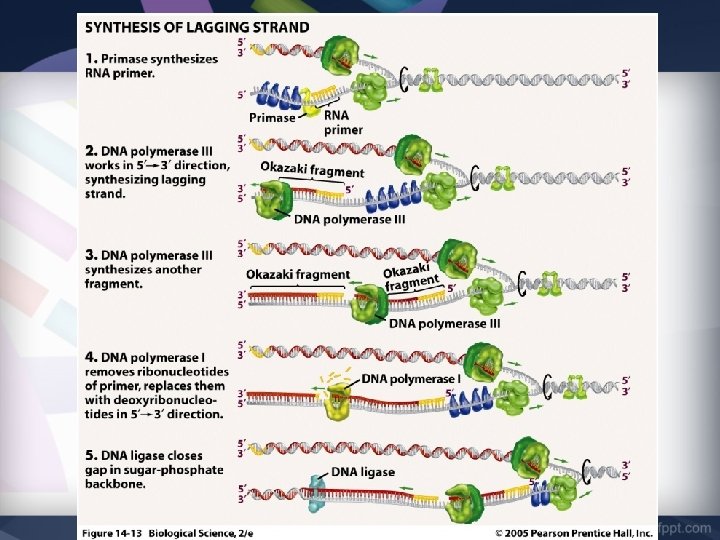
Step 2: Adding RNA Primer • DNA nucleotides cannot bond to “empty” space, so an RNA Primer is used. • Primase is the enzyme responsible for adding the RNA Primer

Step 2: Adding RNA Primer 1. Primase attaches a short sequence of complementary RNA nucleotides to the original strand. 2. Primase adds one RNA primer at the Point of Origin on the Leading Strand (3’ – 5’). 3. Primase has to add multiple RNA Primers on the Lagging Strand (5’ – 3’).

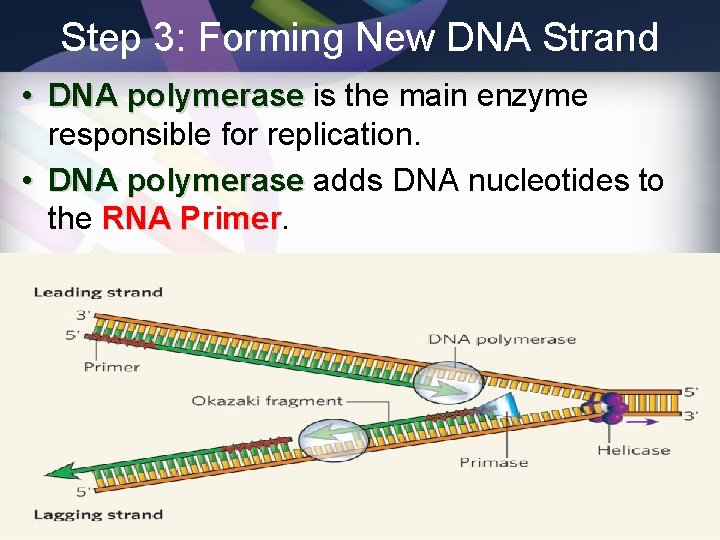
Step 3: Forming New DNA Strand • DNA polymerase is the main enzyme responsible for replication. • DNA polymerase adds DNA nucleotides to the RNA Primer

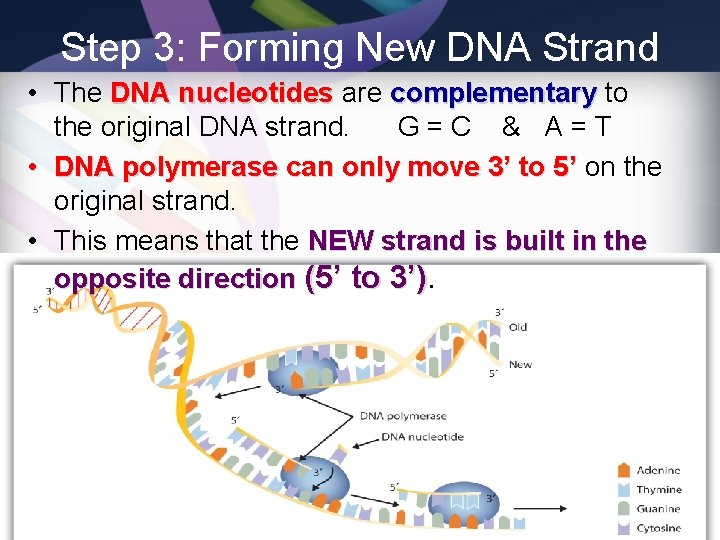
Step 3: Forming New DNA Strand • The DNA nucleotides are complementary to the original DNA strand. G=C & A=T • DNA polymerase can only move 3’ to 5’ on the original strand. • This means that the NEW strand is built in the opposite direction (5’ to 3’)

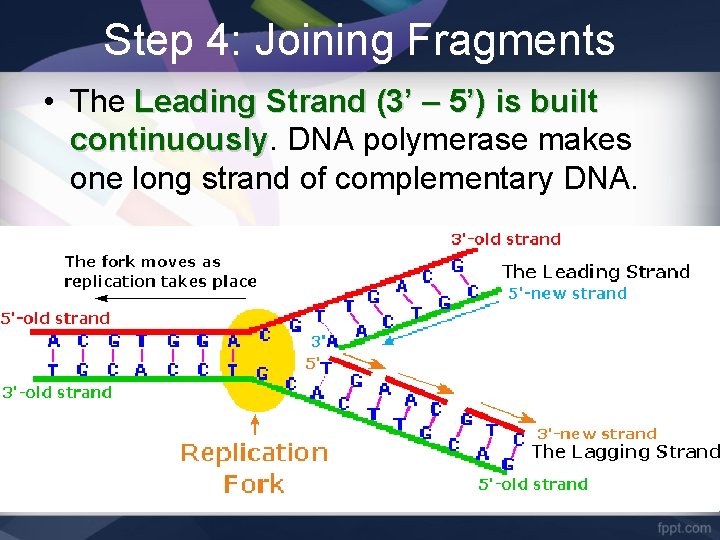
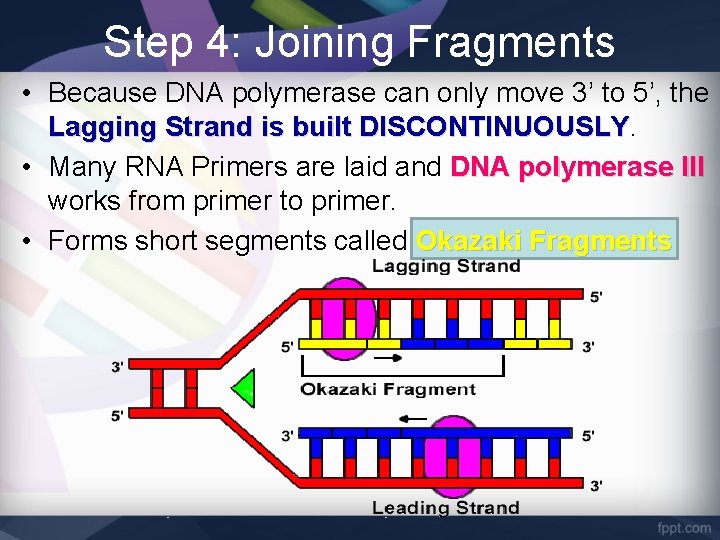
Step 4: Joining Fragments • The Leading Strand (3’ – 5’) is built continuously DNA polymerase makes one long strand of complementary DNA.

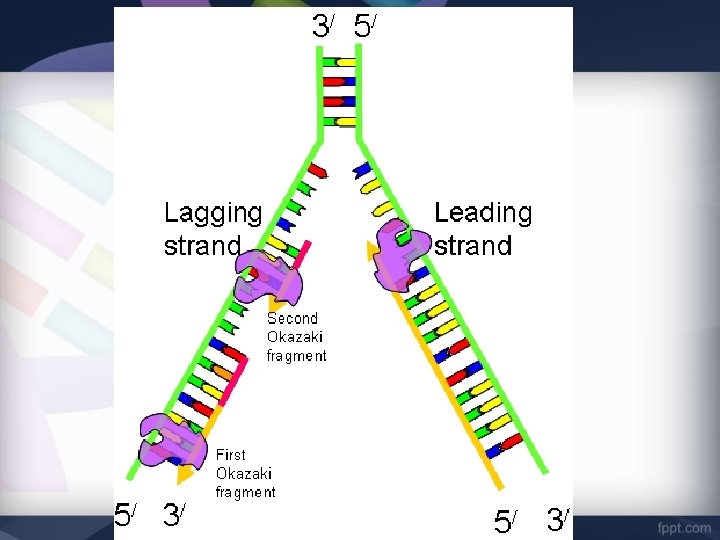
Step 4: Joining Fragments • Because DNA polymerase can only move 3’ to 5’, the Lagging Strand is built DISCONTINUOUSLY • Many RNA Primers are laid and DNA polymerase III works from primer to primer. • Forms short segments called Okazaki Fragments


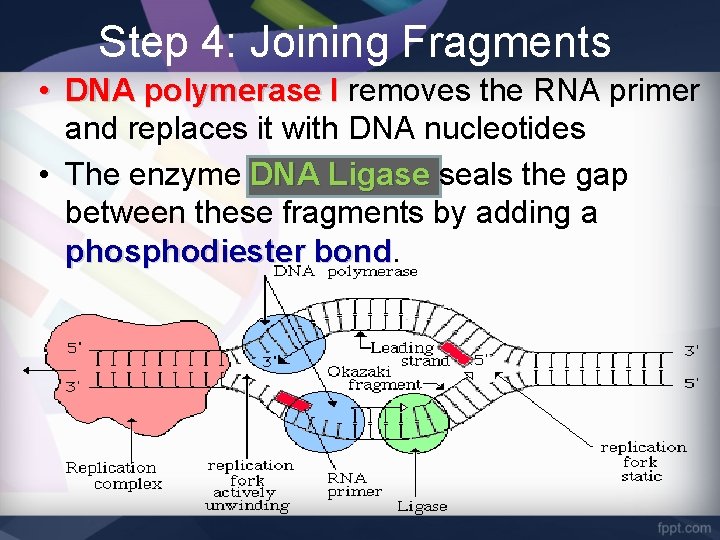
Step 4: Joining Fragments • DNA polymerase I removes the RNA primer and replaces it with DNA nucleotides • The enzyme DNA Ligase seals the gap between these fragments by adding a phosphodiester bond



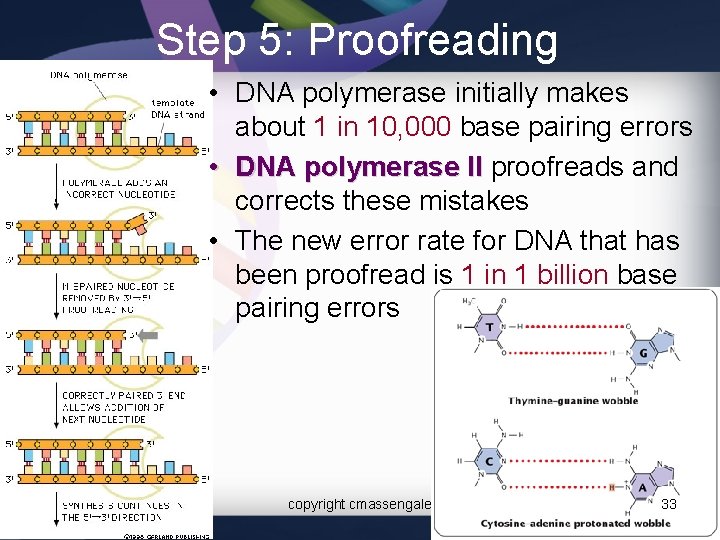
Step 5: Proofreading • DNA polymerase initially makes about 1 in 10, 000 base pairing errors • DNA polymerase II proofreads and corrects these mistakes • The new error rate for DNA that has been proofread is 1 in 1 billion base pairing errors copyright cmassengale 33

DNA replication BIG IDEA Base Pairing ensures that when DNA is replicated, you get 2 identical copies of DNA each with ONE NEW STRAND and ONE ORIGINAL STRAND!