DNA microarrays Affymetrix chips 25 mers 20 per


- Slides: 41

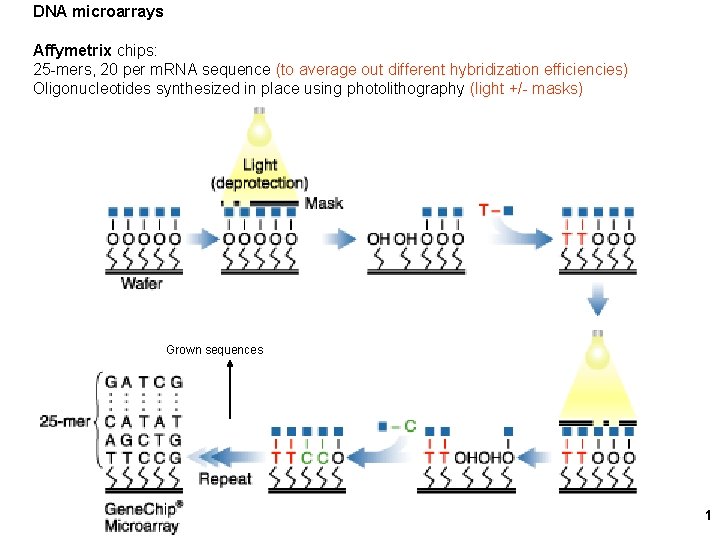
DNA microarrays Affymetrix chips: 25 -mers, 20 per m. RNA sequence (to average out different hybridization efficiencies) Oligonucleotides synthesized in place using photolithography (light +/- masks) Grown sequences 1

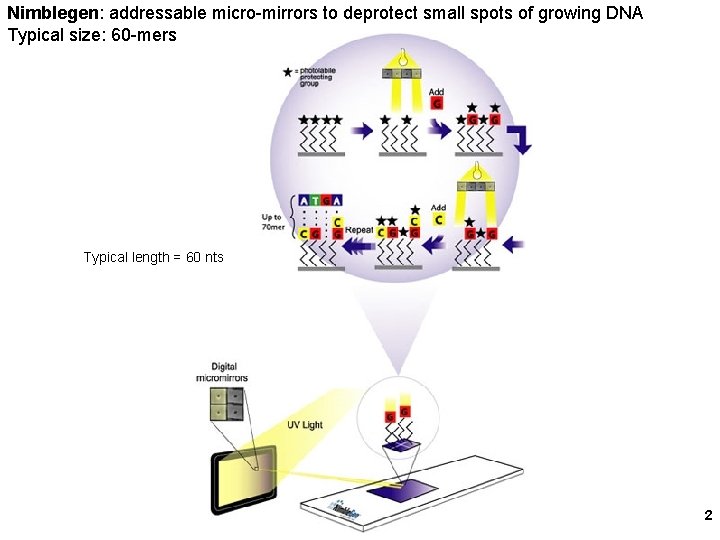
Nimblegen: addressable micro-mirrors to deprotect small spots of growing DNA Typical size: 60 -mers Typical length = 60 nts 2

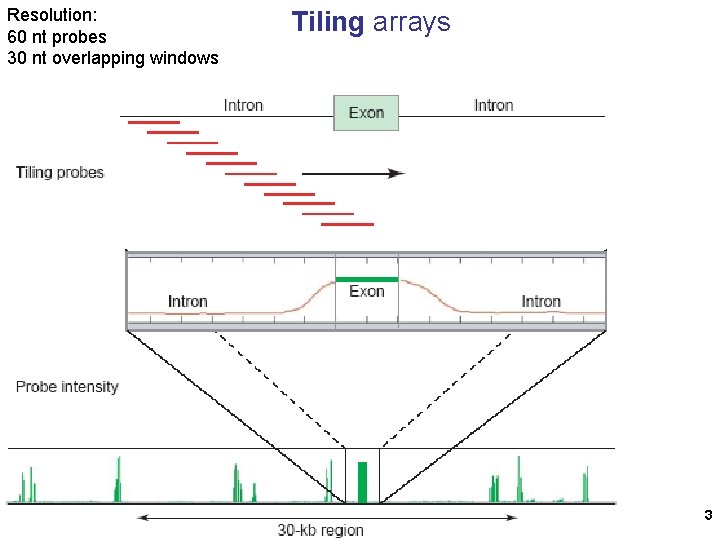
Resolution: 60 nt probes 30 nt overlapping windows Tiling arrays 3

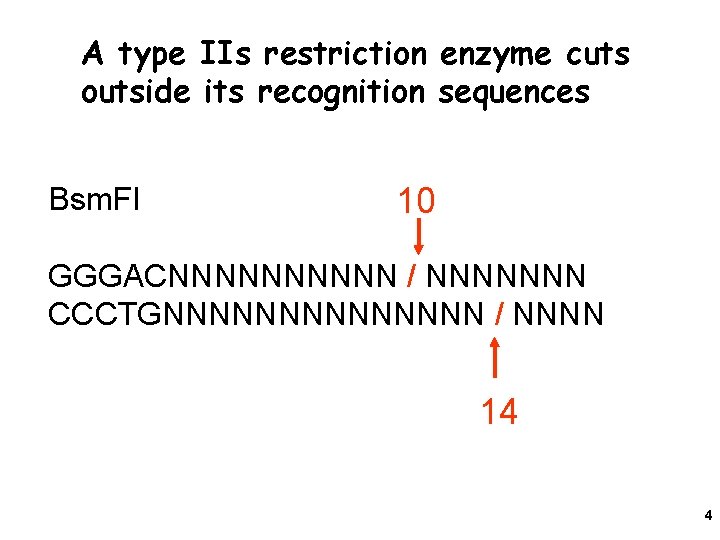
A type IIs restriction enzyme cuts outside its recognition sequences Bsm. FI 10 GGGACNNNNN / NNNNNNN CCCTGNNNNNNN / NNNN 14 4

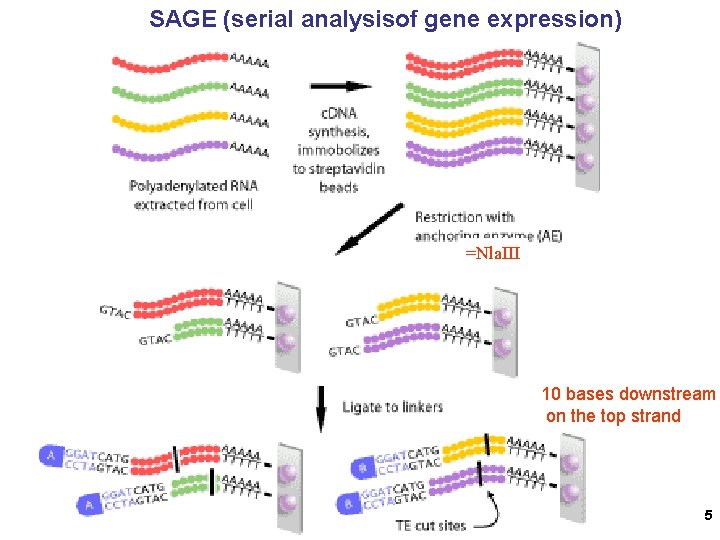
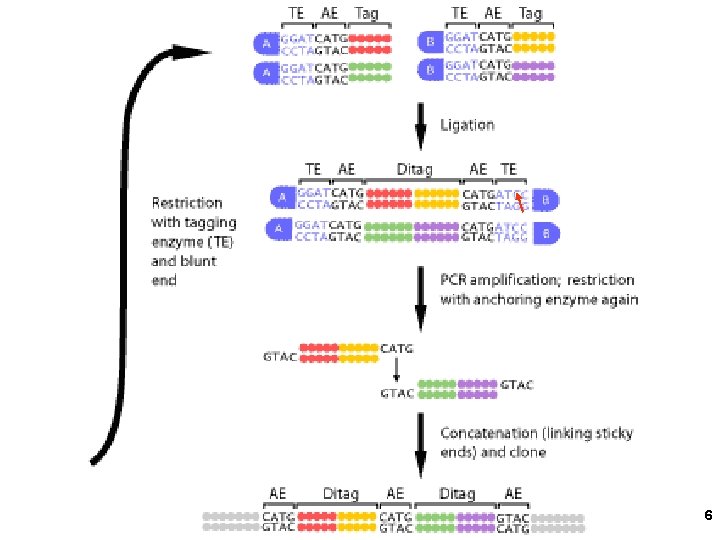
SAGE (serial analysisof gene expression) =Nla. III 10 bases downstream on the top strand 5

6

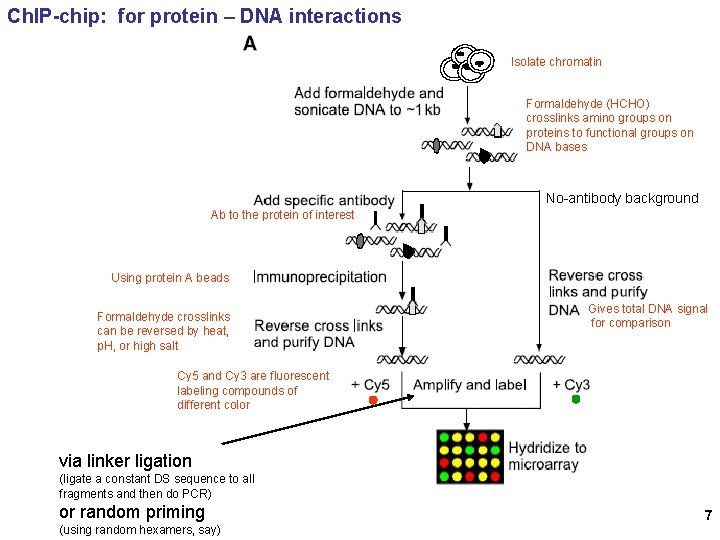
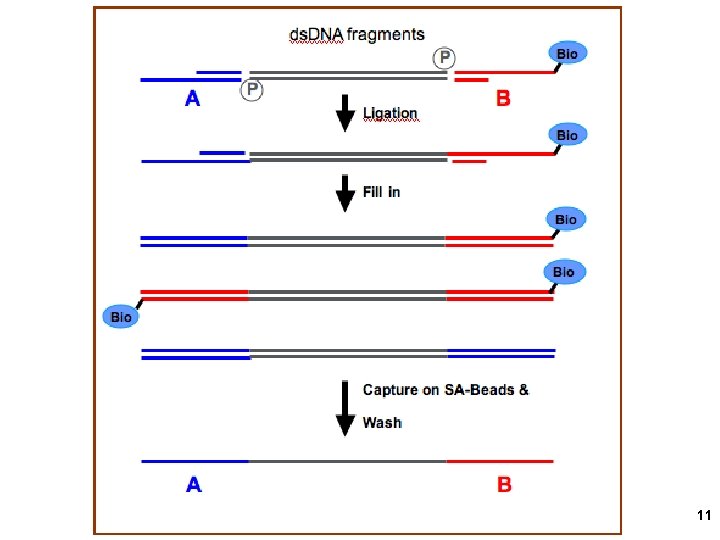
Ch. IP-chip: for protein – DNA interactions Isolate chromatin Formaldehyde (HCHO) crosslinks amino groups on proteins to functional groups on DNA bases No-antibody background Ab to the protein of interest Using protein A beads Formaldehyde crosslinks can be reversed by heat, p. H, or high salt Gives total DNA signal for comparison Cy 5 and Cy 3 are fluorescent labeling compounds of different color via linker ligation (ligate a constant DS sequence to all fragments and then do PCR) or random priming (using random hexamers, say) 7

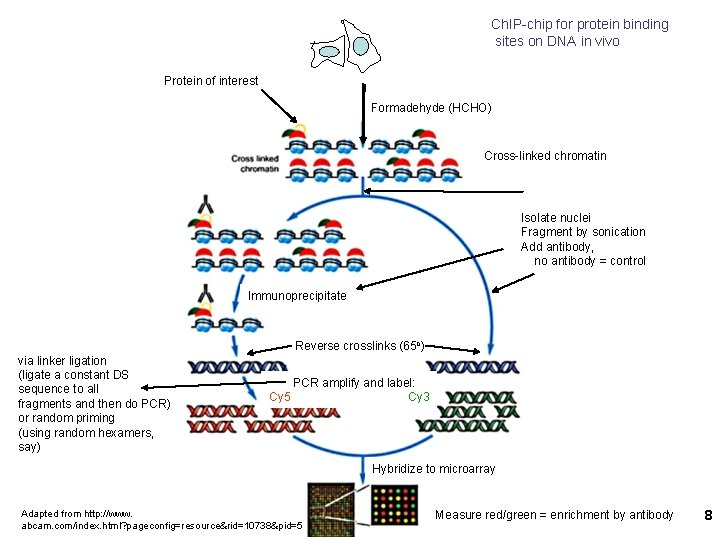
Ch. IP-chip for protein binding sites on DNA in vivo Protein of interest Formadehyde (HCHO) Cross-linked chromatin Isolate nuclei Fragment by sonication Add antibody, no antibody = control Immunoprecipitate Reverse crosslinks (65 o) via linker ligation (ligate a constant DS sequence to all fragments and then do PCR) or random priming (using random hexamers, say) PCR amplify and label: Cy 5 Cy 3 Hybridize to microarray Adapted from http: //www. abcam. com/index. html? pageconfig=resource&rid=10738&pid=5 Measure red/green = enrichment by antibody 8

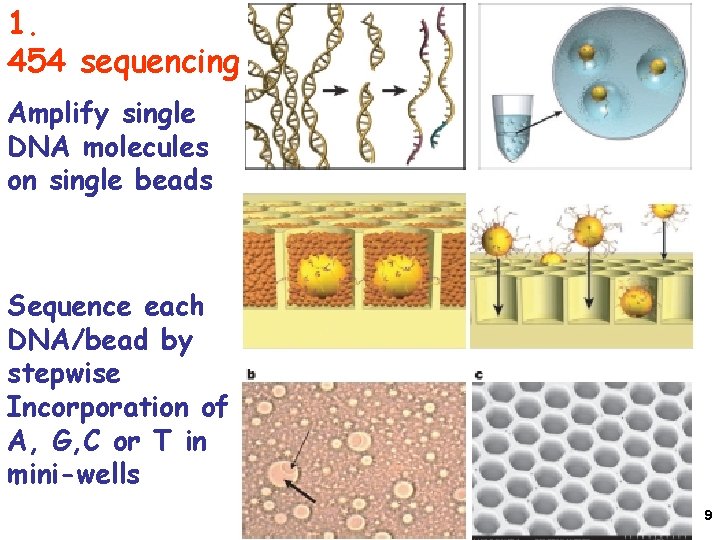
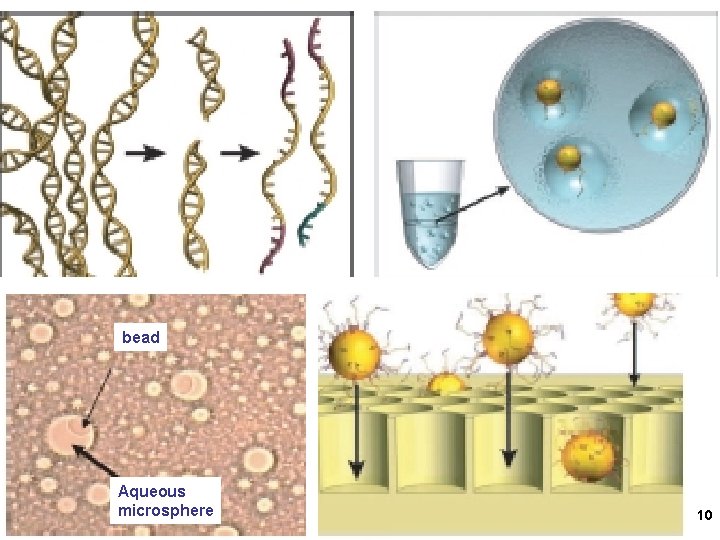
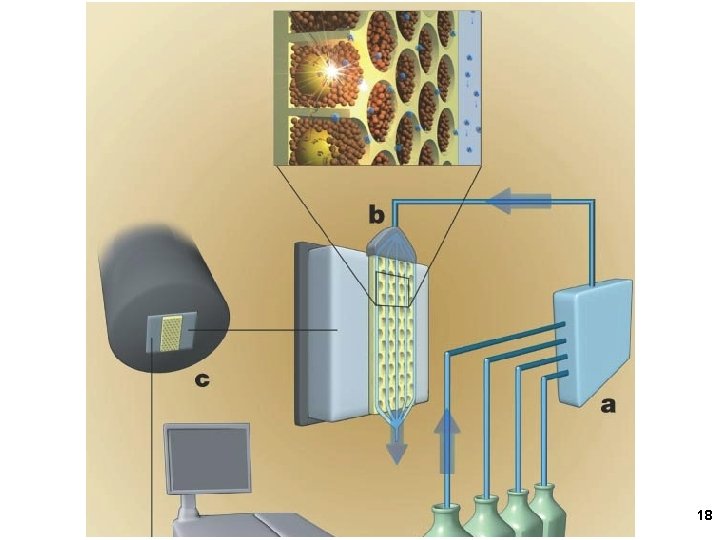
1. 454 sequencing Amplify single DNA molecules on single beads Sequence each DNA/bead by stepwise Incorporation of A, G, C or T in mini-wells 9

bead Aqueous microsphere 10

11

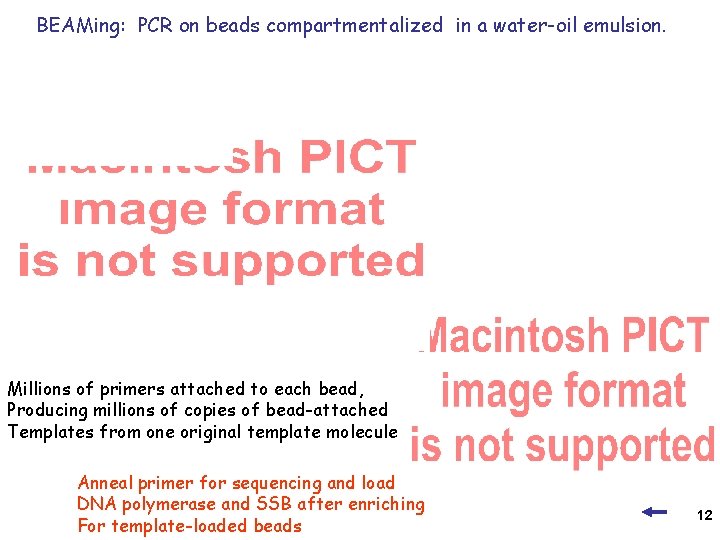
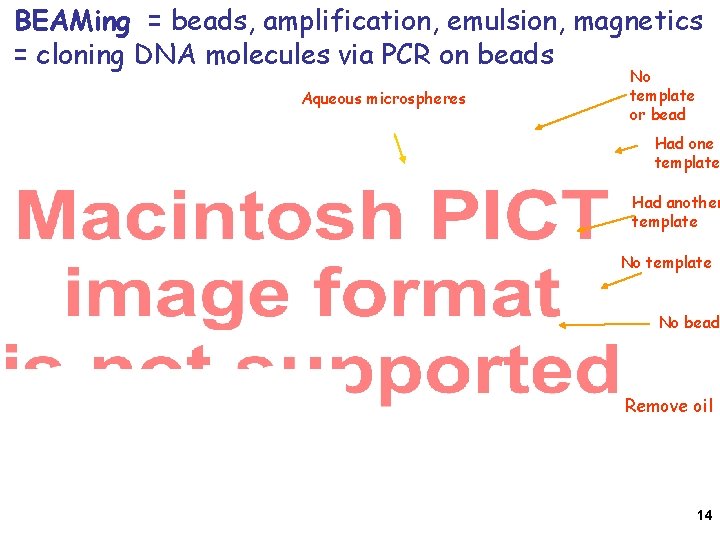
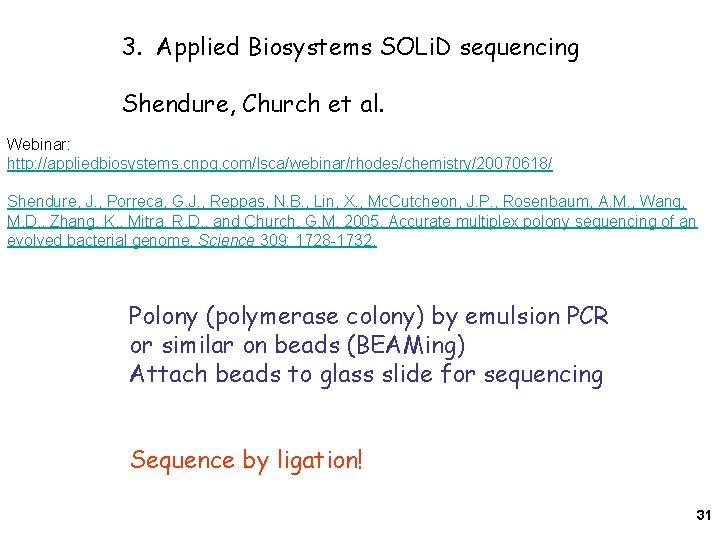
BEAMing: PCR on beads compartmentalized in a water-oil emulsion. Millions of primers attached to each bead, Producing millions of copies of bead-attached Templates from one original template molecule Anneal primer for sequencing and load DNA polymerase and SSB after enriching For template-loaded beads 12

Attached oligomers were pre-labeld red or green, then mixed and emulsified See single beads in aqueous microspheres in oil. 13

BEAMing = beads, amplification, emulsion, magnetics = cloning DNA molecules via PCR on beads Aqueous microspheres No template or bead Had one template Had another template No bead Remove oil 14

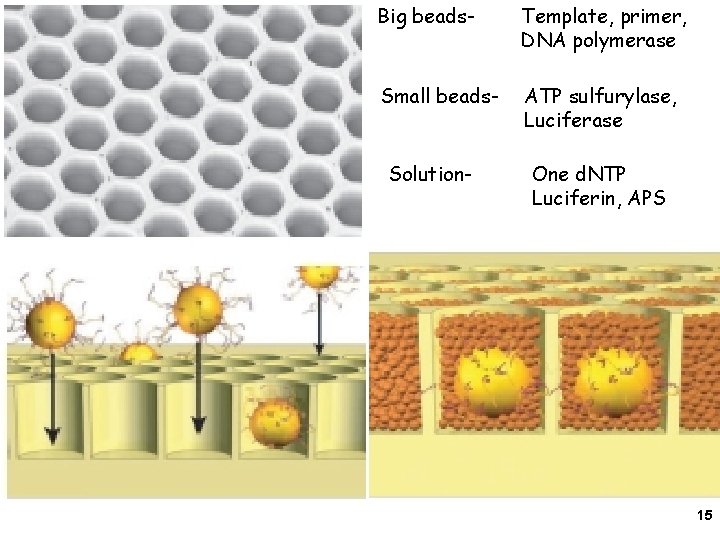
Big beads- Template, primer, DNA polymerase Small beads- ATP sulfurylase, Luciferase Solution- One d. NTP Luciferin, APS 15

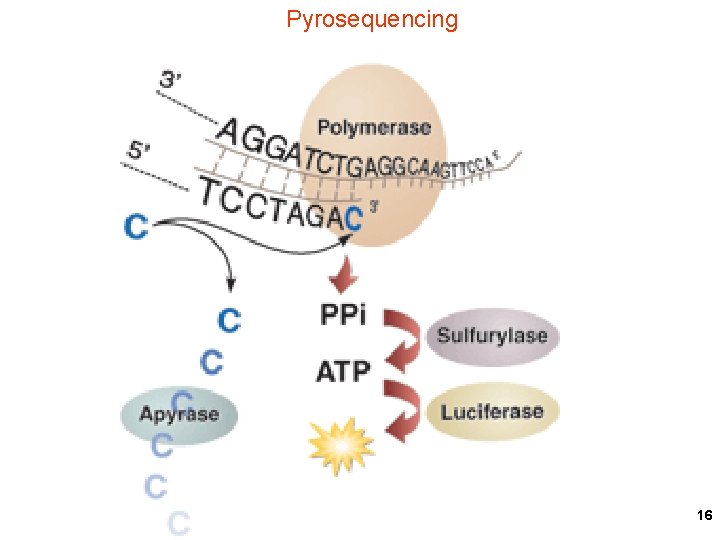
Pyrosequencing 16

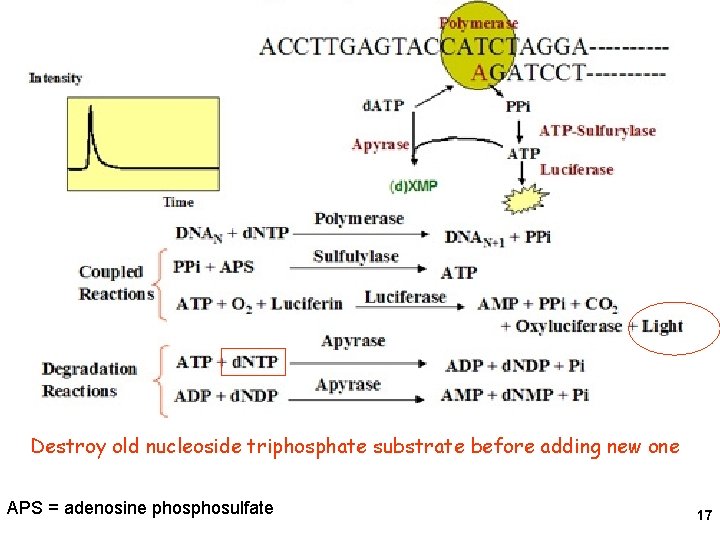
Destroy old nucleoside triphosphate substrate before adding new one APS = adenosine phosulfate 17

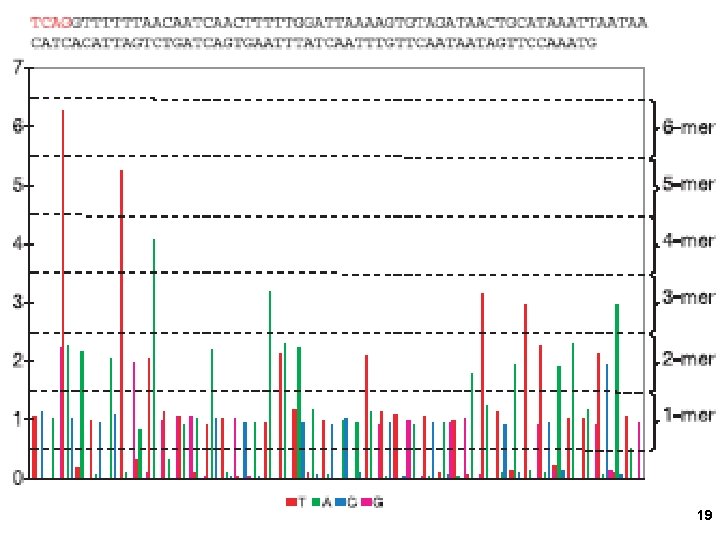
18

19

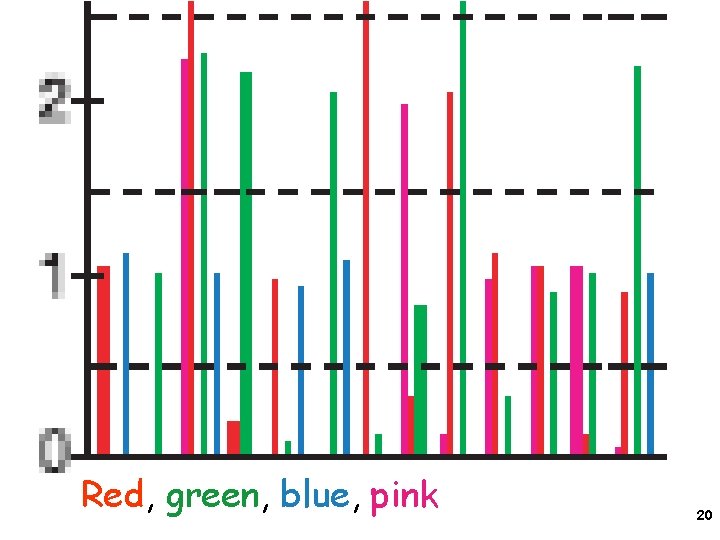
Red, green, blue, pink 20

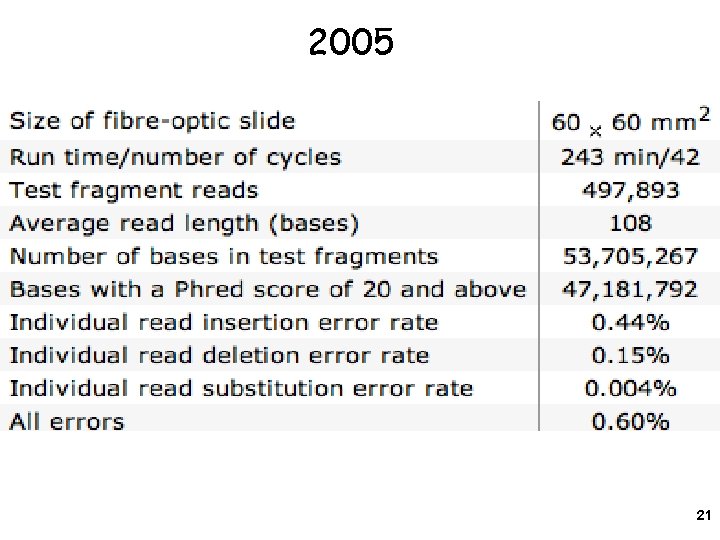
2005 21

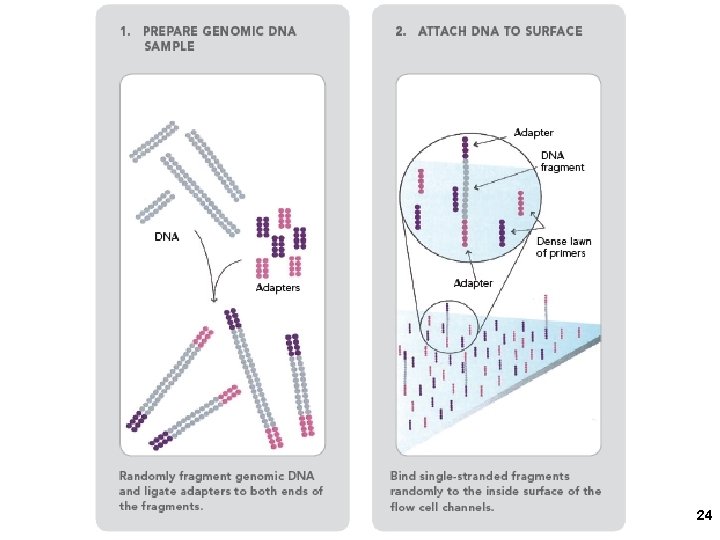
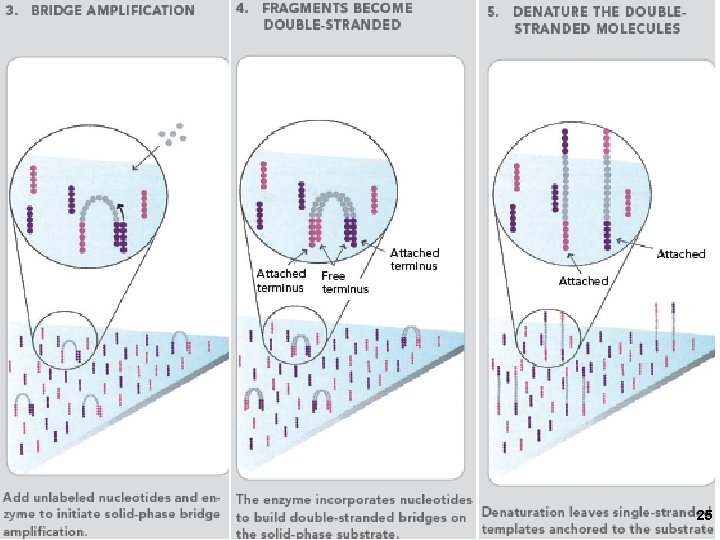
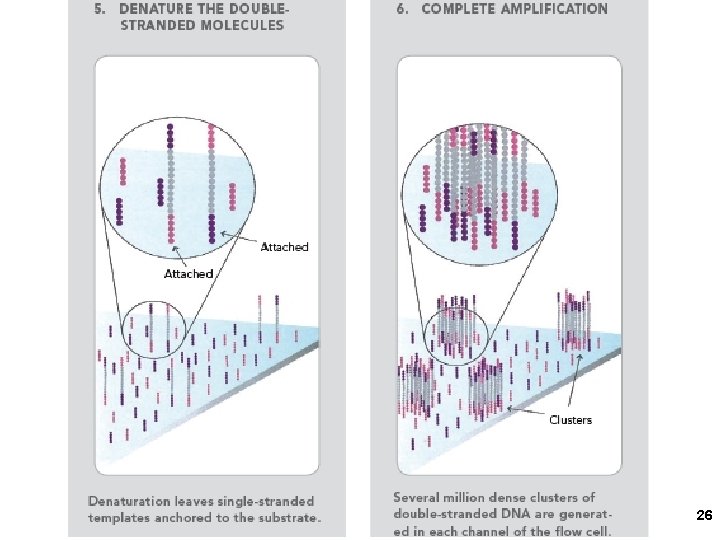
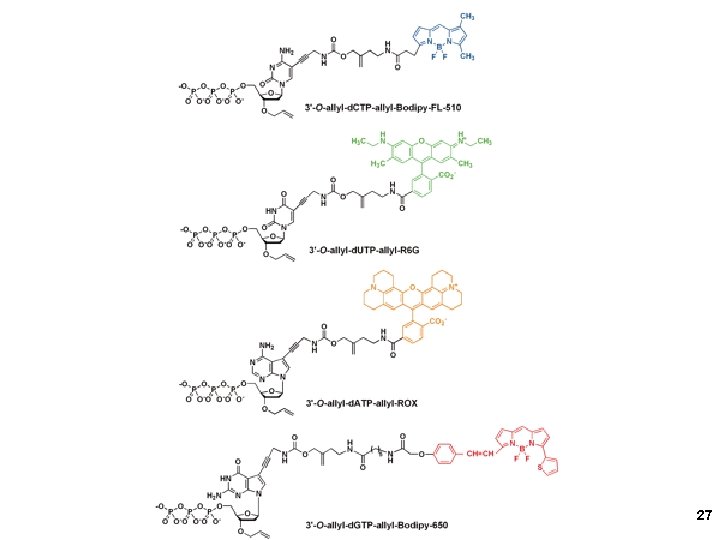
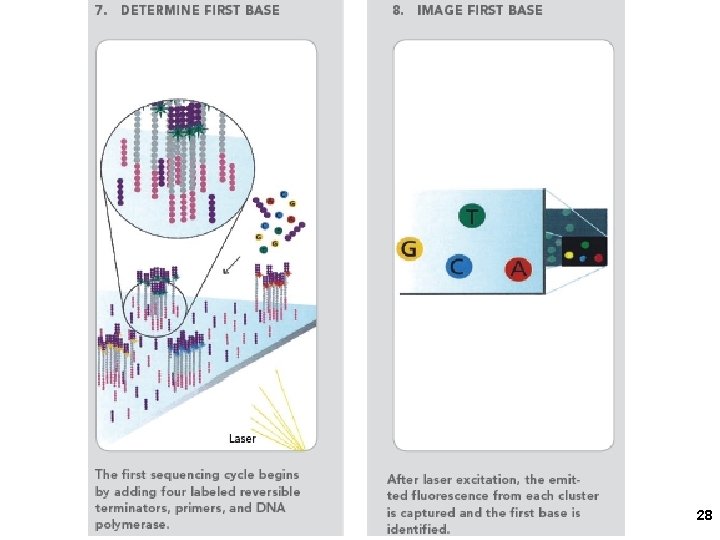
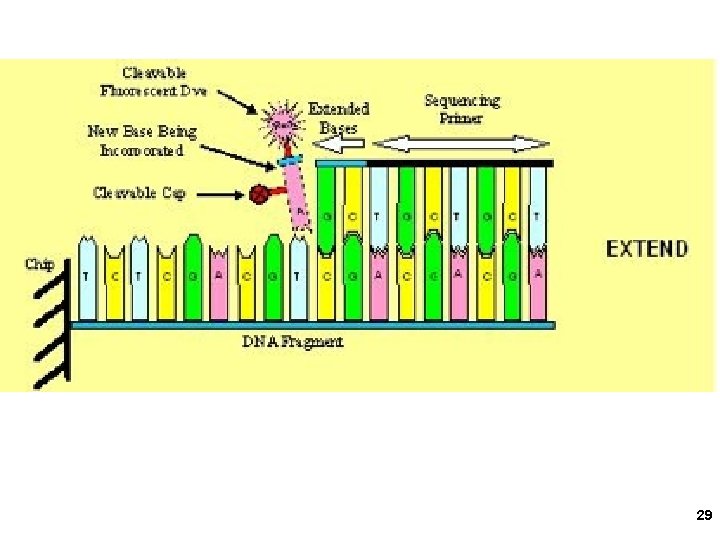
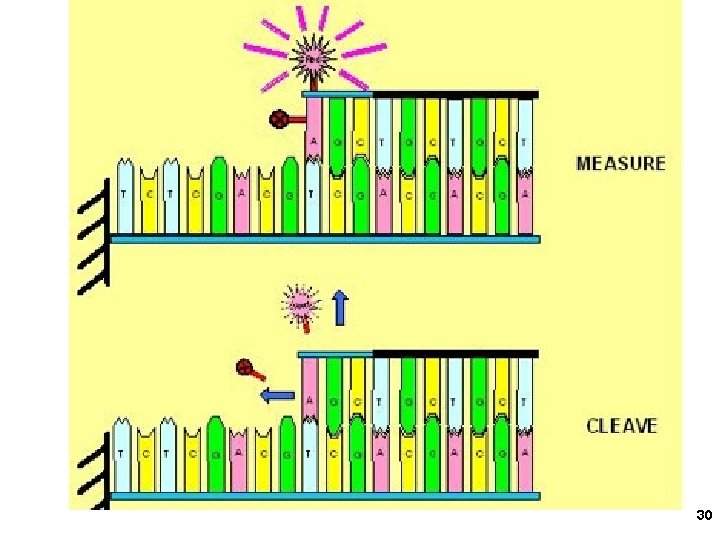
2. Solexa/Illumina sequencing Intelligent Bio-Systems (Jue, Turro… Columbia) Amplification in situ on glass surface of flow cell (PCR that keeps different DNAs separate- “micro-cloning” Sequencing with reversible fluorescent terminator d. NTPs (one nucleotide at a time) 22

Solexa-Illumina 23

24

25

26

27

28

29

30

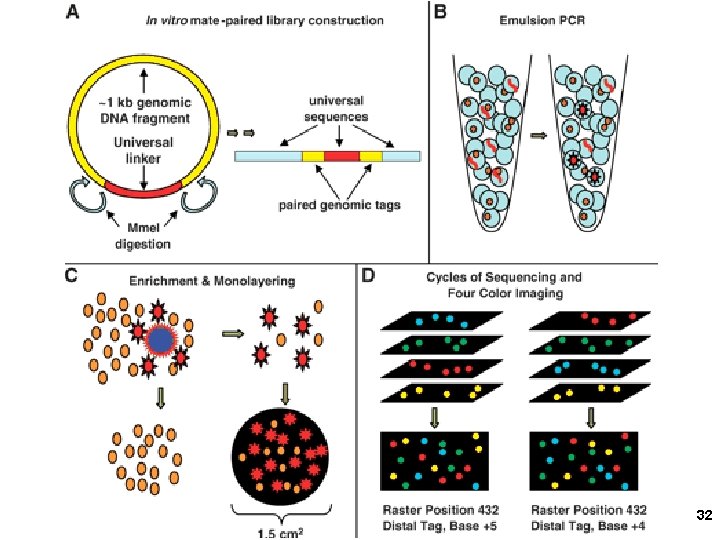
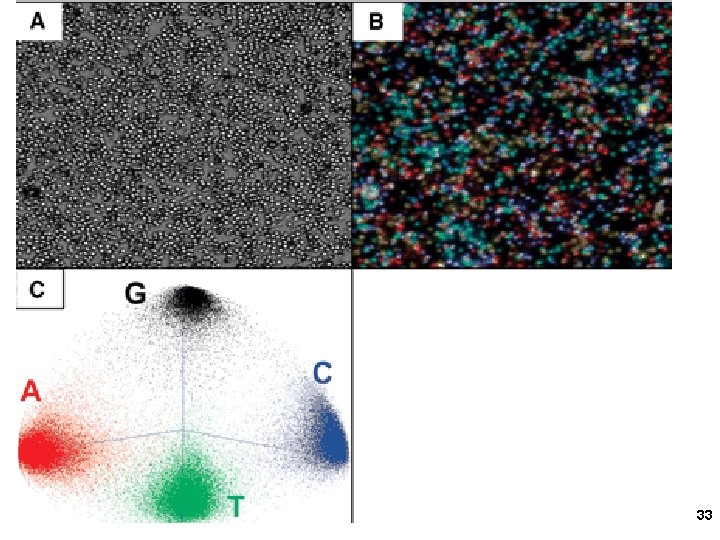
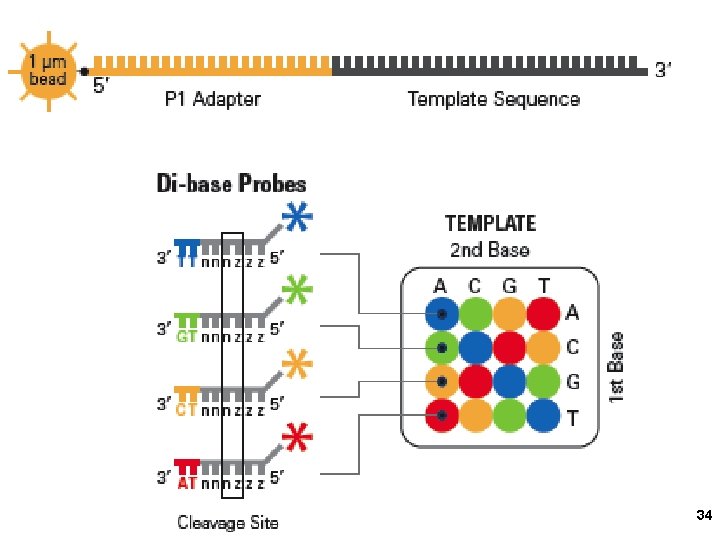
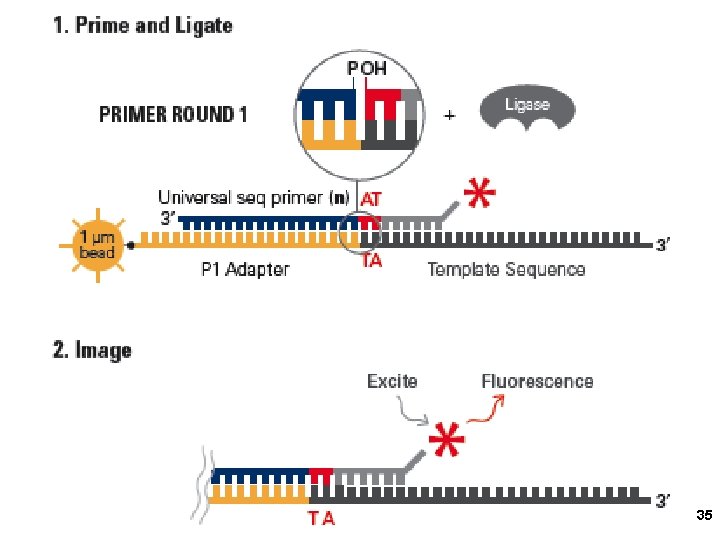
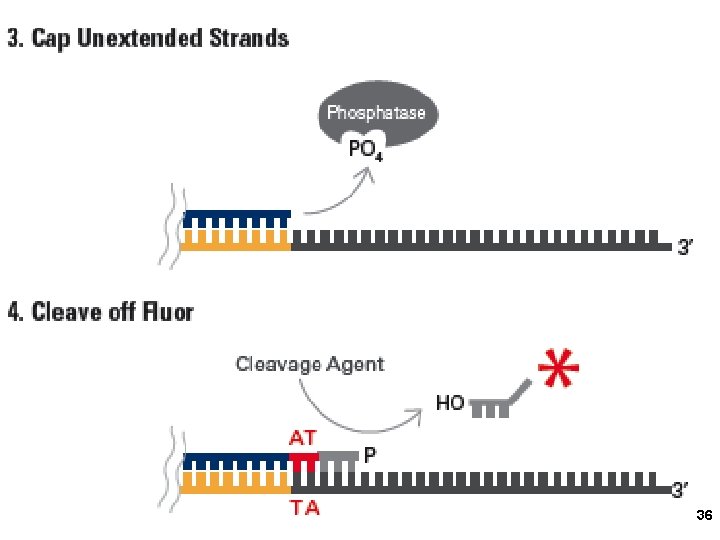
3. Applied Biosystems SOLi. D sequencing Shendure, Church et al. Webinar: http: //appliedbiosystems. cnpg. com/lsca/webinar/rhodes/chemistry/20070618/ Shendure, J. , Porreca, G. J. , Reppas, N. B. , Lin, X. , Mc. Cutcheon, J. P. , Rosenbaum, A. M. , Wang, M. D. , Zhang, K. , Mitra, R. D. , and Church, G. M. 2005. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309: 1728 -1732. Polony (polymerase colony) by emulsion PCR or similar on beads (BEAMing) Attach beads to glass slide for sequencing Sequence by ligation! 31

32

33

34

35

36

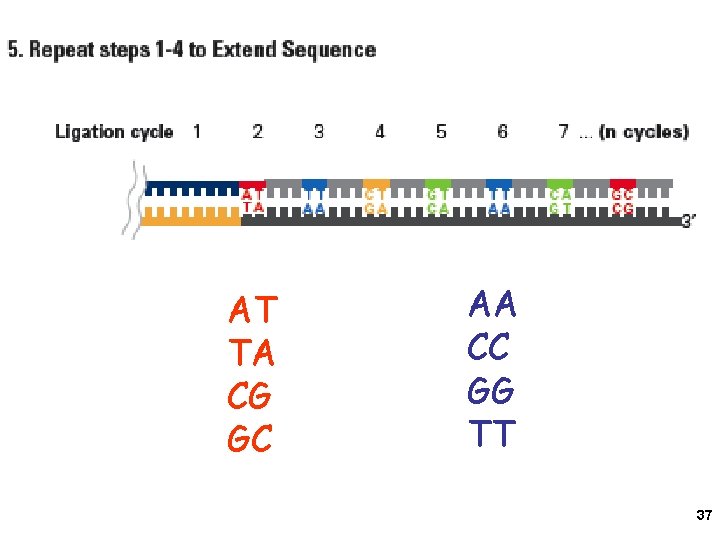
AT TA CG GC AA CC GG TT 37

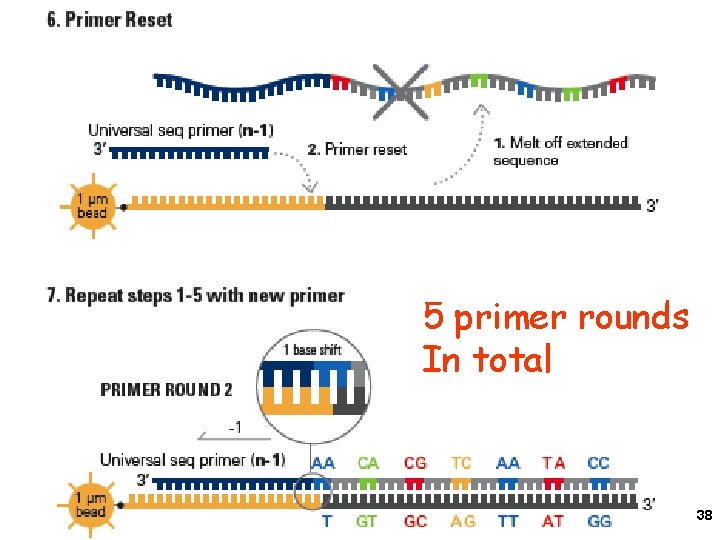
5 primer rounds In total 38

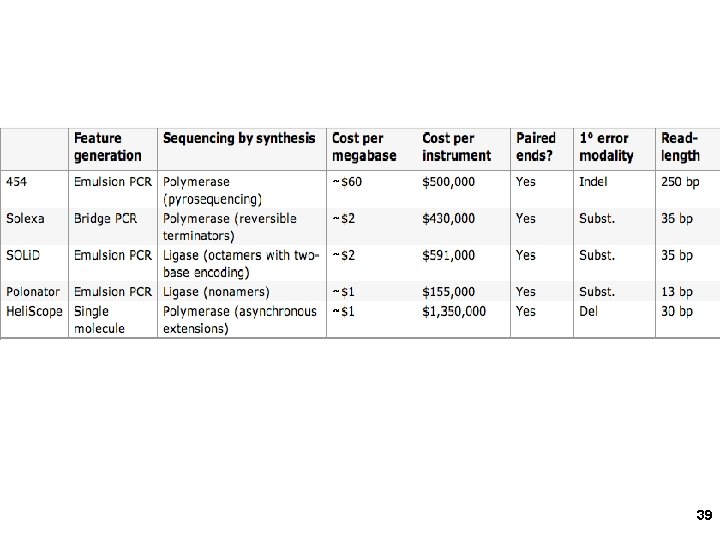
39

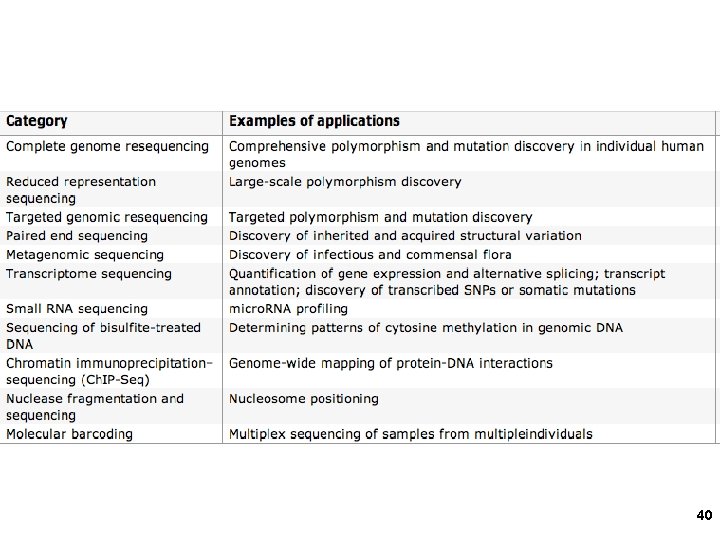
40

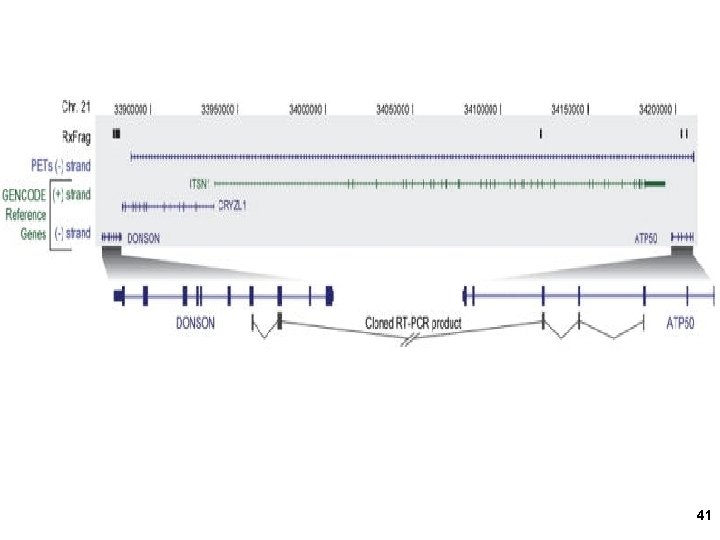
41