Derisking Offshore Oil Exploration with Lipidomics G Todd













- Slides: 28

Derisking Offshore Oil Exploration with Lipidomics G. Todd Ventura

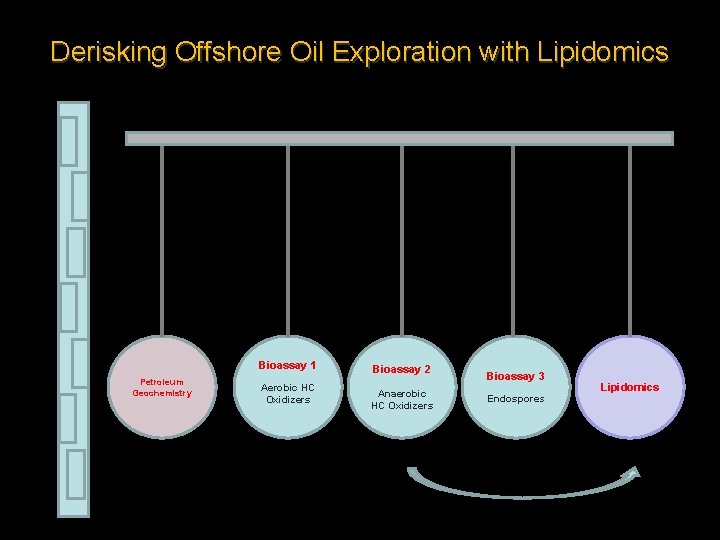
Derisking Offshore Oil Exploration with Lipidomics Bioassay 1 Petroleum Geochemistry Aerobic HC Oxidizers Bioassay 2 Anaerobic HC Oxidizers Bioassay 3 Endospores Lipidomics

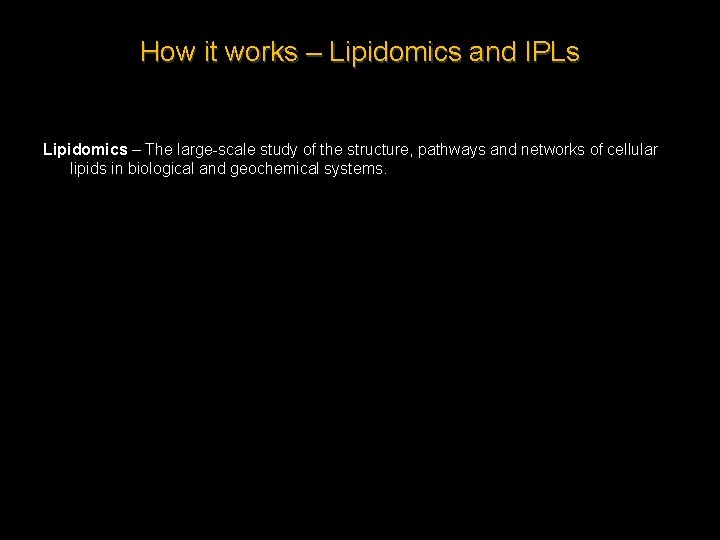
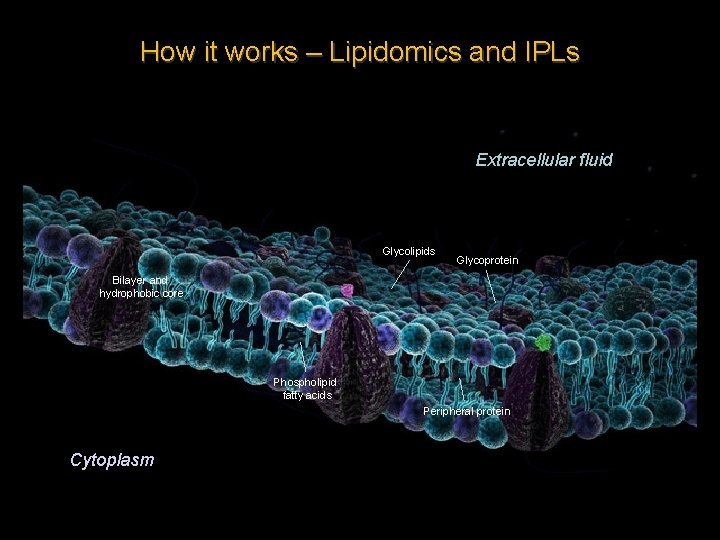
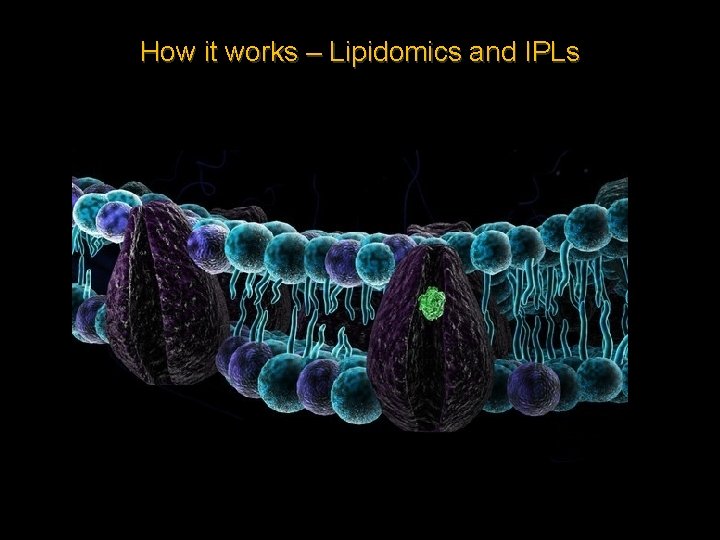
How it works – Lipidomics and IPLs Lipidomics – The large-scale study of the structure, pathways and networks of cellular lipids in biological and geochemical systems.

How it works – Lipidomics and IPLs Extracellular fluid Glycolipids Glycoprotein Bilayer and hydrophobic core Phospholipid fatty acids Peripheral protein Cytoplasm

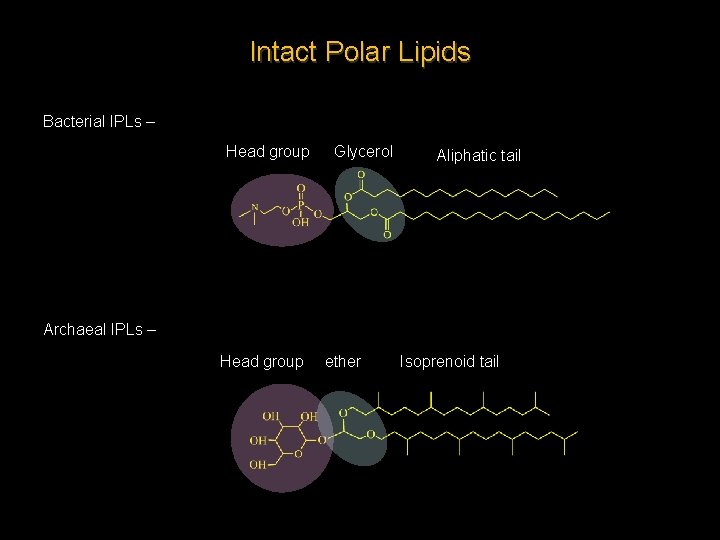
Intact Polar Lipids Bacterial IPLs – Head group Glycerol Aliphatic tail Archaeal IPLs – Head group ether Isoprenoid tail

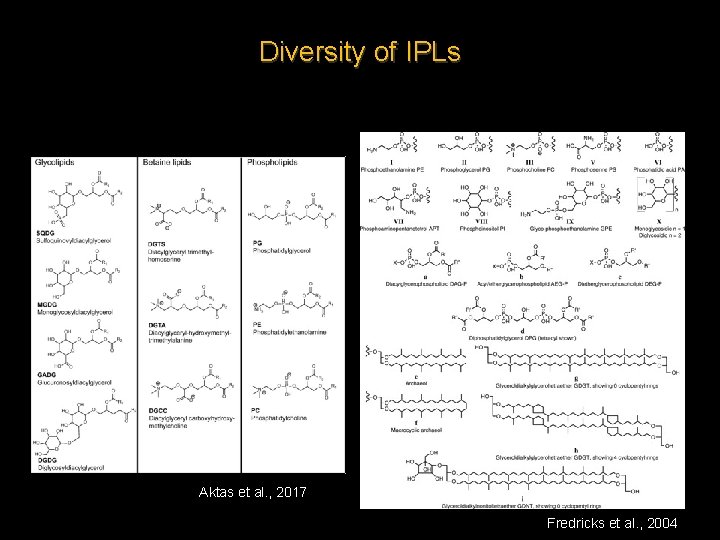
Diversity of IPLs Aktas et al. , 2017 Fredricks et al. , 2004

Other Lipids of Consideration • Fossil Lipids: • Core lipids • Hydrocarbon biomarkers

Research Strategy • Proof of concept • Validation

Research Strategy – Proof of Concept Dr. Carl Peters Mitacs Postdoctoral Fellow Carl has collected frozen piston core samples being stored at Bedford Institute of Oceanography (BIO).

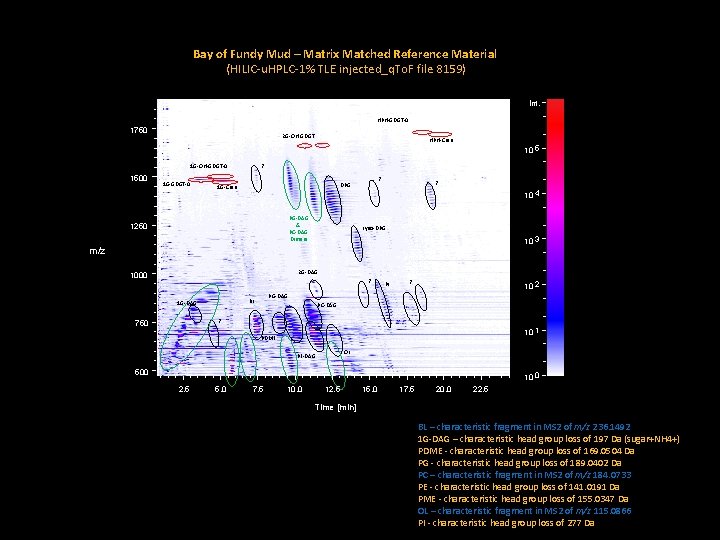
Research Strategy – Proof of Concept Matrix matched reference material – Bay of Fundy estuary mud Total Lipid Extract (TLE) recovery: Number of Repeats = 6

Bay of Fundy Mud – Matrix Matched Reference Material (HILIC-u. HPLC-1% TLE injected_q. To. F file 8159) Int. HPH-GDGT-0 1750 2 G-OH-GDGT 1 G-GDGT-0 10 5 ? 1 G-OH-GDGT-0 1500 HPH-Cren ? DPG 1 G-Cren PG-DAG & PC-DAG Dimers 1250 ? 10 4 Lyso-DPG 10 3 m/z 2 G-DAG 1000 ? ? 10 2 PG-DAG BL 1 G-DAG PI PC-DAG ? 750 10 1 PDME OL PE-DAG 500 10 0 2. 5 5. 0 7. 5 10. 0 12. 5 15. 0 17. 5 20. 0 22. 5 Time [min] BL – characteristic fragment in MS 2 of m/z 236. 1492 1 G-DAG – characteristic head group loss of 197 Da (sugar+NH 4+) PDME - characteristic head group loss of 169. 0504 Da PG - characteristic head group loss of 189. 0402 Da PC – characteristic fragment in MS 2 of m/z 184. 0733 PE - characteristic head group loss of 141. 0191 Da PME - characteristic head group loss of 155. 0347 Da OL – characteristic fragment in MS 2 of m/z 115. 0866 PI - characteristic head group loss of 277 Da

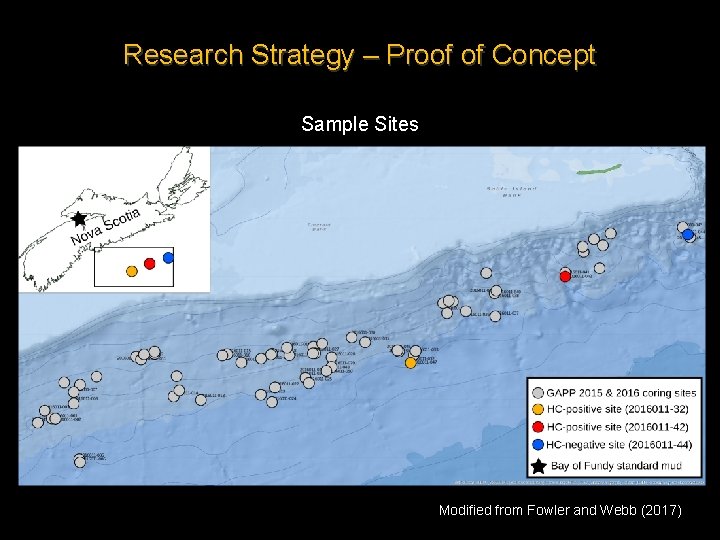
Research Strategy – Proof of Concept Sample Sites Modified from Fowler and Webb (2017)

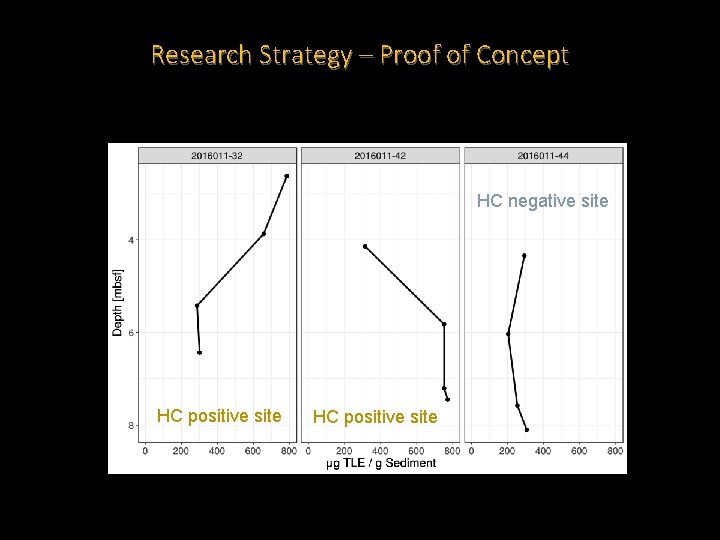
Research Strategy – Proof of Concept HC negative site HC positive site

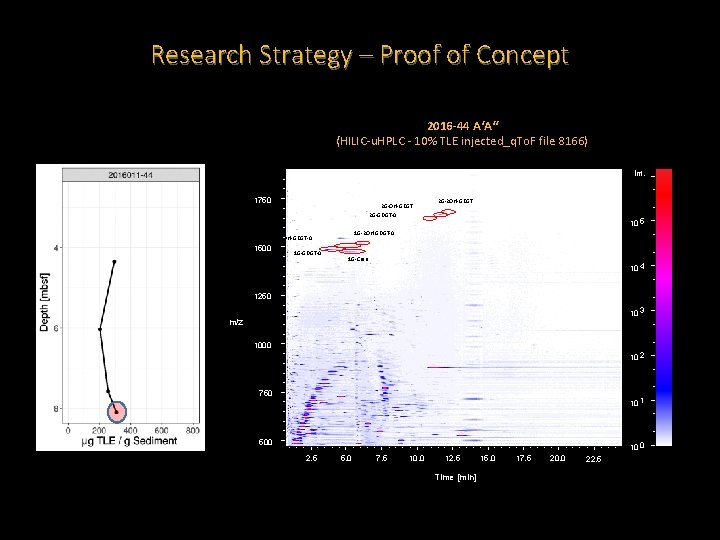
Research Strategy – Proof of Concept 2016 -44 A‘A‘‘ (HILIC-u. HPLC - 10% TLE injected_q. To. F file 8166) Int. 1750 2 G-OH-GDGT 2 G-GDGT-0 10 5 1 G-2 OH-GDGT-0 1 G-OH-GDGT-0 1500 2 G-2 OH-GDGT 1 G-Cren 10 4 1250 10 3 m/z 1000 10 2 750 10 1 500 10 0 2. 5 5. 0 7. 5 10. 0 12. 5 Time [min] 15. 0 17. 5 20. 0 22. 5

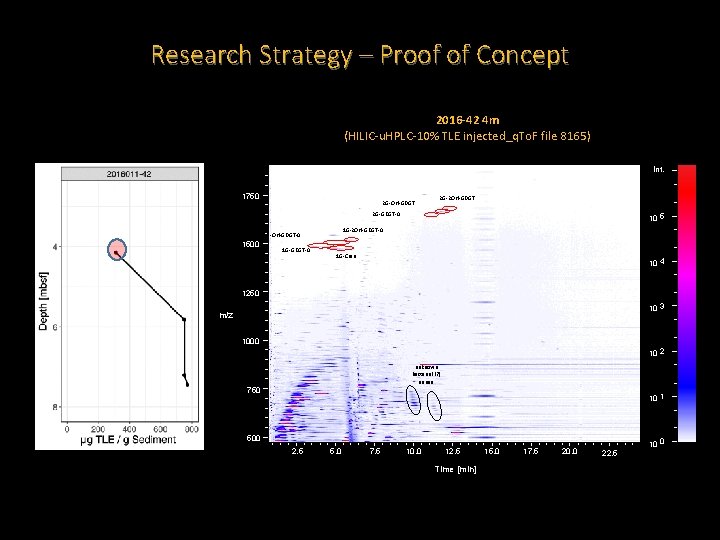
Research Strategy – Proof of Concept 2016 -42 4 m (HILIC-u. HPLC-10% TLE injected_q. To. F file 8165) Int. 1750 2 G-OH-GDGT 2 G-2 OH-GDGT 2 G-GDGT-0 1 G-2 OH-GDGT-0 1 G-OH-GDGT-0 1500 1 G-GDGT-0 10 5 1 G-Cren 10 4 1250 10 3 m/z 1000 10 2 unknown bacterial (? ) series 750 10 1 500 2. 5 5. 0 7. 5 10. 0 12. 5 Time [min] 15. 0 17. 5 20. 0 10 0 22. 5

Research Strategy – Proof of Concept • Reproducible extraction method that is yielding lipids. • Extracts from HC positive Scotian shelf piston cores appear to be consistently greater than for a HC negative site. • The Scotian Shelf sediments have IPLs that are much less diverse than what we see for our reference standard. • Both HC positive and negative sites have Archaeal IPLs. • So far, the HC positive site also contains what are likely to be Bacterial IPLs.

Research Strategy – Proof of Concept • Reproducible extraction method that is yielding lipids. • Extracts from HC positive Scotian shelf piston cores appear to be consistently greater than for a HC negative site. • The Scotian Shelf sediments have IPLs that are much less diverse than what we see for our reference standard. • Both HC positive and negative sites have Archaeal IPLs. • So far, the HC positive site also contains what are likely to be Bacterial IPLs. Research Strategy – Validation

Research Strategy - Validation Dr. Casey Hubert and his group have conducted three sampling seasons. 2015 – Where, CCGS Hudson 2016 – Where, CCGS Hudson 2017 – Sydney Basin, RV Coriolis

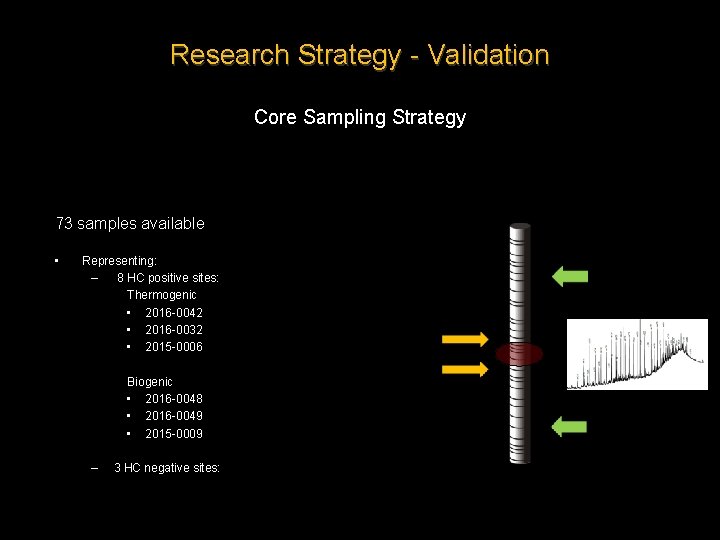
Research Strategy - Validation Core Sampling Strategy 73 samples available • Representing: – 8 HC positive sites: Thermogenic • 2016 -0042 • 2016 -0032 • 2015 -0006 Biogenic • 2016 -0048 • 2016 -0049 • 2015 -0009 – 3 HC negative sites:

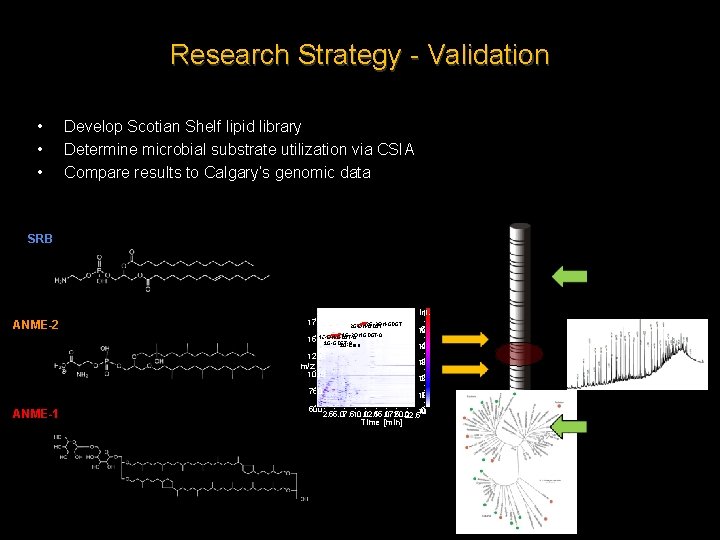
Research Strategy - Validation • • • Develop Scotian Shelf lipid library Determine microbial substrate utilization via CSIA Compare results to Calgary’s genomic data SRB Int. ANME-2 1750 2 G-2 OH-GDGT 2 G-OH-GDGT 1 G-2 OH-GDGT-0 1 G-OH-GDGT-0 1500 1 G-GDGT-0 1 G-Cren 4 10 1250 3 10 m/z ANME-1 5 10 1000 2 10 750 1 10 500 0 2. 55. 07. 510. 0 12. 5 15. 0 17. 5 20. 0 22. 510 Time [min]

Conclusions • We believe we have the right strategy and support network to evaluate the use of IPLs as IHIs for offshore exploration in Nova Scotia. • Stay tuned for more!

Acknowledgements Collaborators: Casey Hubert, Jayne Rattray, Julius Lipp, Florence Schubotz, and Kai Uwe-Hinrichs. Funding Sources: - Canadian Research Chair - Canadian Foundation for Innovation – JELF - Offshore Exploration Research Association (Halifax, Nova Scotia, Canada) - Department of Energy Nova Scotia - Mitacs - SMU FGSR

Thank you

How it works – Lipidomics and IPLs

Derisking Offshore Oil Exploration with Lipidomics • We are working to generate lipid profiles extracted from piston core sediments collected at known seep sites to target subsurface living microbial communities that are actively degrading petroleum-based hydrocarbons. • This project aims to compare the sensitivity of lipidomics with that of the University of Calgary’s bioassay technology. • The lipid assignments for hydrocarbon oxidation are equally to be compared with the petroleum geochemistry data produced by APT Ltd. Canada.

Microbial Hydrocarbon Oxidation Contingent is the hypothesis working is that microbes are proliferating in seep sediments by anaerobically oxidizing petroleum forming hydrocarbons How does this look:

Microbial Hydrocarbon Oxidation

 Classes of lipids
Classes of lipids Upstream oil exploration
Upstream oil exploration Well production
Well production Preparation of emulsion
Preparation of emulsion I am looking for a new challenge
I am looking for a new challenge London standard drilling barge form
London standard drilling barge form K line offshore as
K line offshore as Iii offshore advisors
Iii offshore advisors Structural dynamics of deep water offshore wind turbines
Structural dynamics of deep water offshore wind turbines Offshore drilling
Offshore drilling Offshore employment intermediaries
Offshore employment intermediaries Offshore drilling
Offshore drilling Offshore drilling
Offshore drilling Offshore pipes
Offshore pipes What is offshore construction
What is offshore construction Offshore graphic design
Offshore graphic design Offshore delivery assessment
Offshore delivery assessment Fracking natural gas
Fracking natural gas Offshore drilling
Offshore drilling Offshore wind accelerator
Offshore wind accelerator Dwmis
Dwmis Tiffany oil rig
Tiffany oil rig Gastropanel
Gastropanel Offshore drilling
Offshore drilling Offshore drilling
Offshore drilling Nyserda offshore wind master plan
Nyserda offshore wind master plan Pepe and alfredo are resting on an offshore raft
Pepe and alfredo are resting on an offshore raft Ata offshore
Ata offshore Bmsb target high risk goods
Bmsb target high risk goods