Course on Functional Analysis Introduction to Functional Analysis








- Slides: 26

Course on Functional Analysis : : : Introduction to Functional Analysis ? Daniel Rico, Ph. D. drico@cnio. es Bioinformatics Unit CNIO

: : : Schedule. 1. 2. 3. 4. Biological (Functional) Databases Threshold-based and threshold free methods Threshold-based example: Fati. GO. Threshold free example 1: Fatis. Scan.

ACKNOWLEDGEMENTS Many of these slides have been taken and adapted from original slides by Fatima Al-Shahrour from Joaquin Dopazo’s group (Babelomics team). We are grateful for the material and for the great tools they have developed!!!!

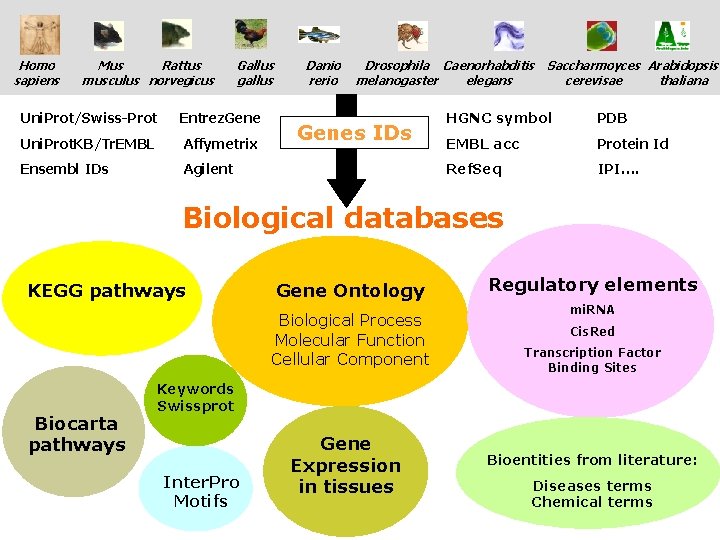
Homo sapiens Mus Rattus musculus norvegicus Gallus gallus Uni. Prot/Swiss-Prot Entrez. Gene Uni. Prot. KB/Tr. EMBL Affymetrix Ensembl IDs Agilent Danio rerio Drosophila Caenorhabditis melanogaster elegans Genes IDs Saccharmoyces Arabidopsis thaliana cerevisae HGNC symbol PDB EMBL acc Protein Id Ref. Seq IPI…. Biological databases KEGG pathways Gene Ontology Biological Process Molecular Function Cellular Component Biocarta pathways Regulatory elements mi. RNA Cis. Red Transcription Factor Binding Sites Keywords Swissprot Inter. Pro Motifs Gene Expression in tissues Bioentities from literature: Diseases terms Chemical terms

Gene Ontology CONSORTIUM http: //www. geneontology. org • The objective of GO is to provide controlled vocabularies for the description of the molecular function, biological process and cellular component of gene products. • These terms are to be used as attributes of gene products by collaborating databases, facilitating uniform queries across them. • The controlled vocabularies of terms are structured

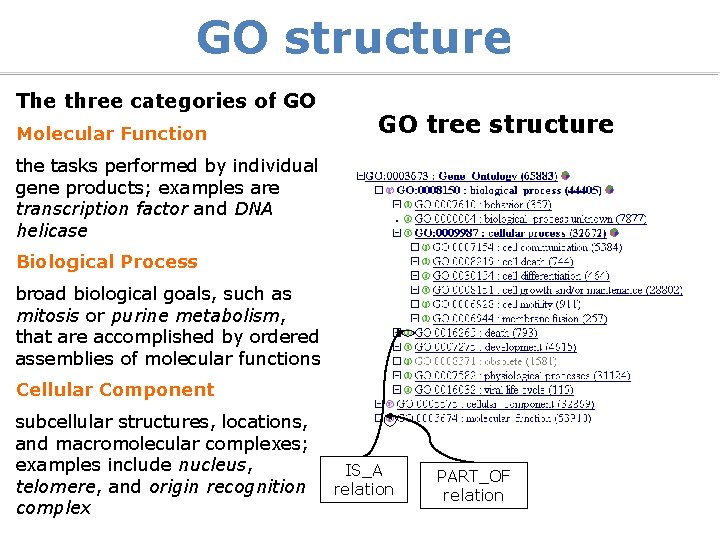
GO structure The three categories of GO Molecular Function GO tree structure the tasks performed by individual gene products; examples are transcription factor and DNA helicase Biological Process broad biological goals, such as mitosis or purine metabolism, that are accomplished by ordered assemblies of molecular functions Cellular Component subcellular structures, locations, and macromolecular complexes; examples include nucleus, telomere, and origin recognition complex IS_A relation PART_OF relation

http: //www. genome. ad. jp/kegg/pathway. html

http: //www. biocarta. com/genes/index. asp

http: //www. reactome. org/

http: //www. pathwaycommons. org

http: //www. whichgenes. org/

http: //www. cisred. org/

: : : Schedule. 1. 2. 3. 4. Biological (Functional) Databases Threshold-based and threshold free methods Threshold-based example: Fati. GO. Threshold free example 1: Fatis. Scan.

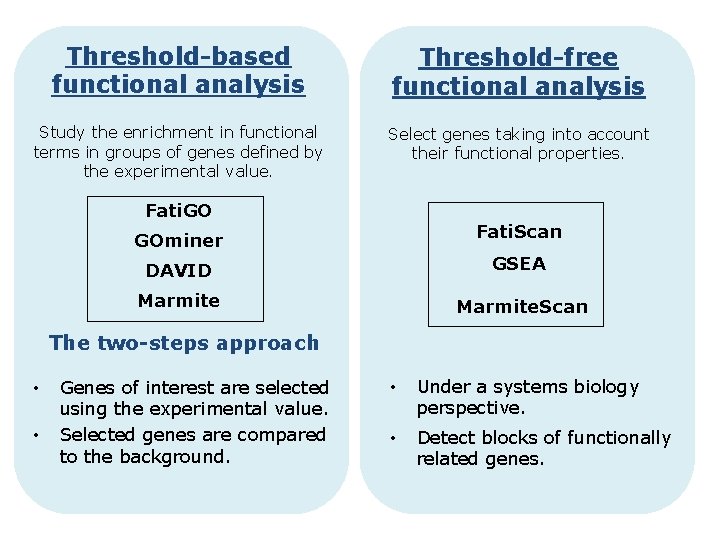
Threshold-based functional analysis Threshold-free functional analysis Study the enrichment in functional terms in groups of genes defined by the experimental value. Select genes taking into account their functional properties. Fati. GO Fati. Scan GOminer DAVID GSEA Marmite. Scan The two-steps approach • • Genes of interest are selected using the experimental value. Selected genes are compared to the background. • Under a systems biology perspective. • Detect blocks of functionally related genes.

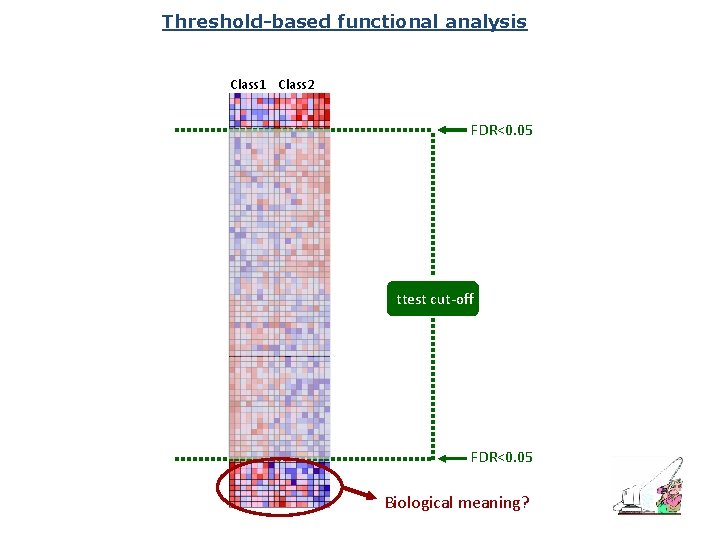
Threshold-based functional analysis Class 1 Class 2 FDR<0. 05 ttest cut-off FDR<0. 05 Biological meaning?

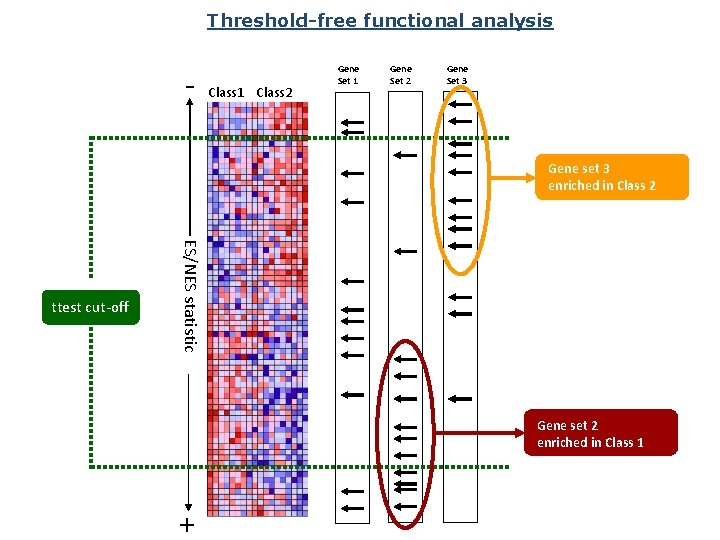
Threshold-free functional analysis - Class 1 Class 2 Gene Set 1 Gene Set 2 Gene Set 3 Gene set 3 enriched in Class 2 ES/NES statistic ttest cut-off Gene set 2 enriched in Class 1 +

: : : Schedule. 1. 2. 3. 4. Biological (Functional) Databases Threshold-based and threshold free methods Threshold-based example: Fati. GO. Threshold free example 1: Fatis. Scan.

http: //babelomics. bioinfo. cipf. es/

: : : How the functional profiling should never be done It is not uncommon to find the following assertion in papers and talks: “then we examined our set of genes selected in this way (whatever) and we discover that 65% of them were related to metabolism, so we can conclude that our experiment activates metabolism genes”. Annotation is not a functional result!!!

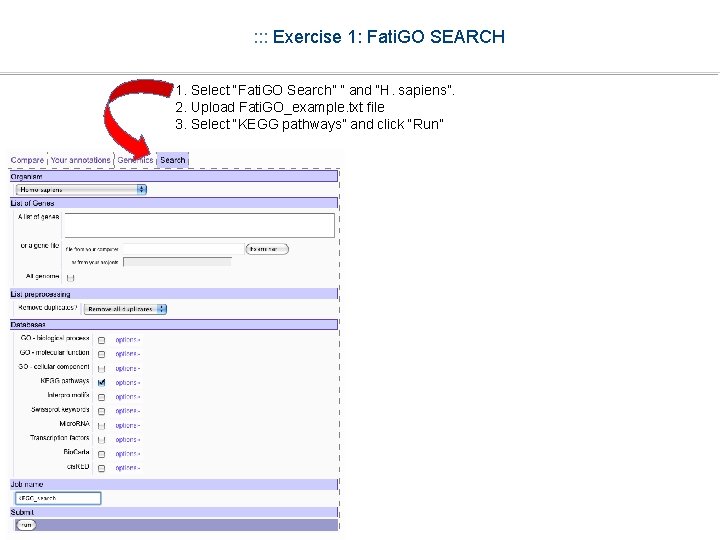
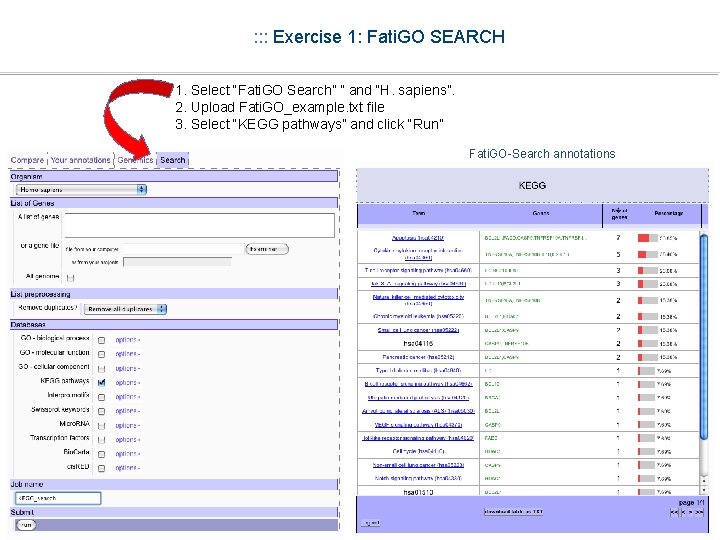
: : : Exercise 1: Fati. GO SEARCH 1. Select “Fati. GO Search” ” and “H. sapiens”. 2. Upload Fati. GO_example. txt file 3. Select “KEGG pathways” and click “Run”

: : : Exercise 1: Fati. GO SEARCH 1. Select “Fati. GO Search” ” and “H. sapiens”. 2. Upload Fati. GO_example. txt file 3. Select “KEGG pathways” and click “Run” Fati. GO-Search annotations

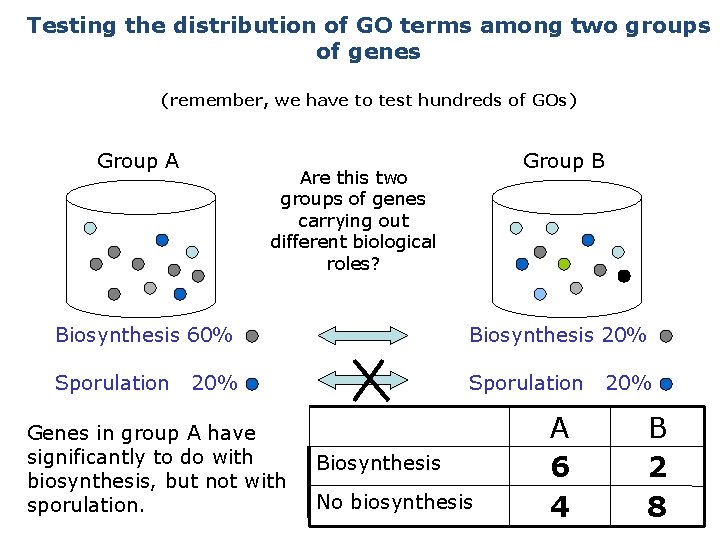
Testing the distribution of GO terms among two groups of genes (remember, we have to test hundreds of GOs) Group A Group B Are this two groups of genes carrying out different biological roles? Biosynthesis 60% Biosynthesis 20% Sporulation 20% Genes in group A have significantly to do with biosynthesis, but not with sporulation. Biosynthesis No biosynthesis A 6 4 20% B 2 8

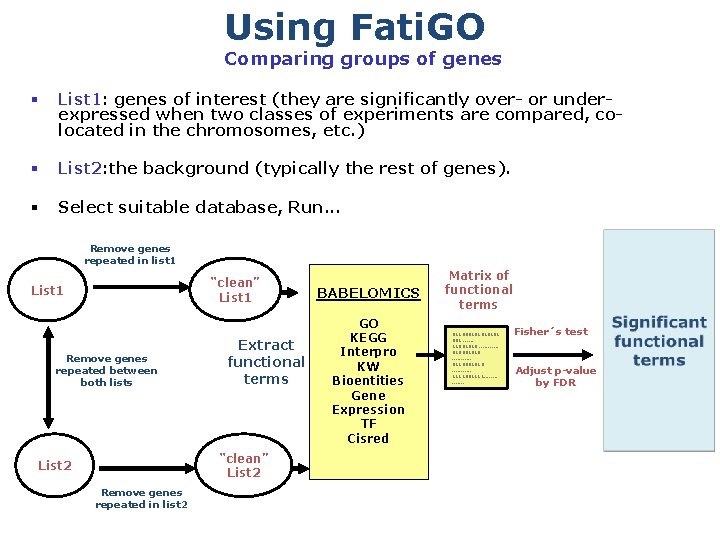
Using Fati. GO Comparing groups of genes List 1: genes of interest (they are significantly over- or underexpressed when two classes of experiments are compared, colocated in the chromosomes, etc. ) List 2: the background (typically the rest of genes). Select suitable database, Run. . . Remove genes repeated in list 1 “clean” List 1 Remove genes repeated between both lists Extract functional terms “clean” List 2 Remove genes repeated in list 2 BABELOMICS GO KEGG Interpro KW Bioentities Gene Expression TF Cisred Matrix of functional terms 01100010101 001. . . 11001010. . . 010001010. . . 0110001010. . . 1111001111. . . . Fisher´s test Adjust p-value by FDR

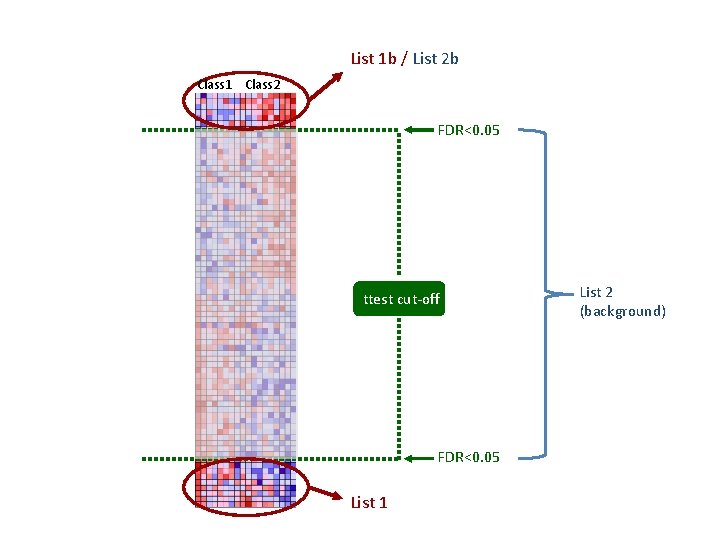
List 1 b / List 2 b Class 1 Class 2 FDR<0. 05 ttest cut-off FDR<0. 05 List 1 List 2 (background)

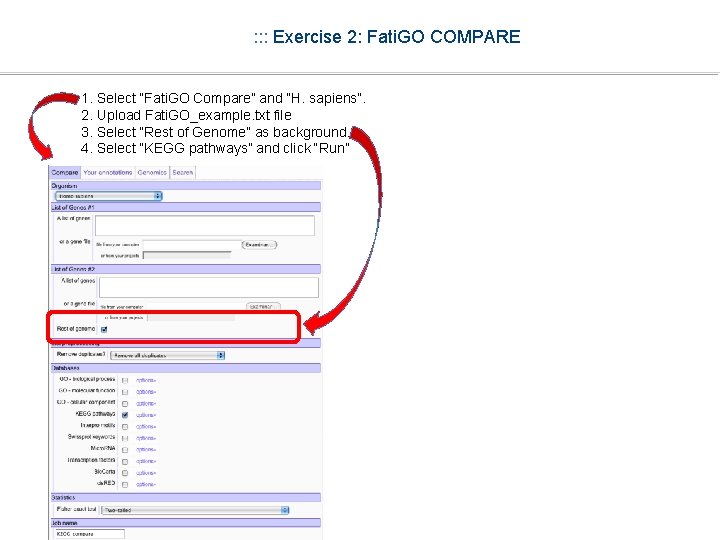
: : : Exercise 2: Fati. GO COMPARE 1. Select “Fati. GO Compare” and “H. sapiens”. 2. Upload Fati. GO_example. txt file 3. Select “Rest of Genome” as background. 4. Select “KEGG pathways” and click “Run”

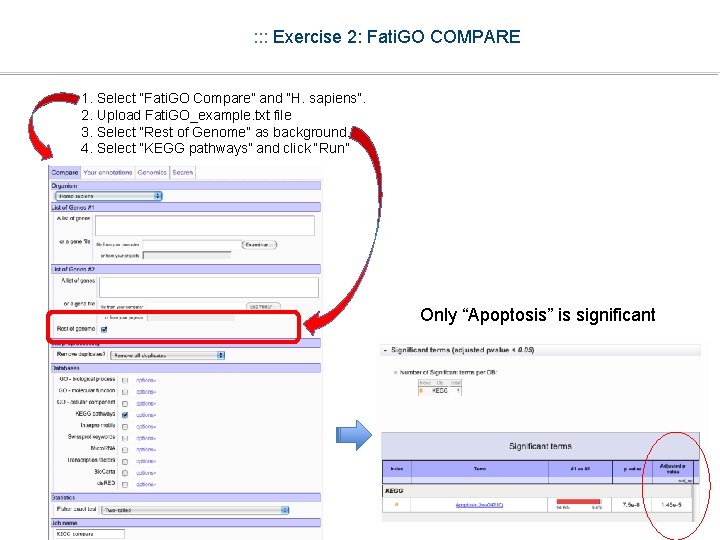
: : : Exercise 2: Fati. GO COMPARE 1. Select “Fati. GO Compare” and “H. sapiens”. 2. Upload Fati. GO_example. txt file 3. Select “Rest of Genome” as background. 4. Select “KEGG pathways” and click “Run” Only “Apoptosis” is significant