Clinical Cytogenetics Disorders of the Autosomes Autosomal Disorders

























- Slides: 43

Clinical Cytogenetics: Disorders of the Autosomes

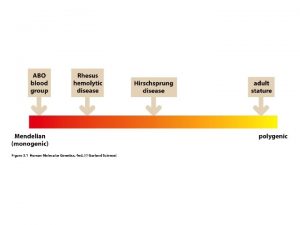
Autosomal Disorders Only 3 well-defined non-mosaic chr disorders compatible with postnatal survival in which there is trisomy for an entire chr. (13, 18, 21) l Each is associated with growth retardation, MR and multiple congenital abnormalities. But each has a distinctive phenotype l Abnormalities determined by extra dosage of the particular genes on the additional chr. l Specific genes on the extra chr. responsible for abnormal phenotypes l

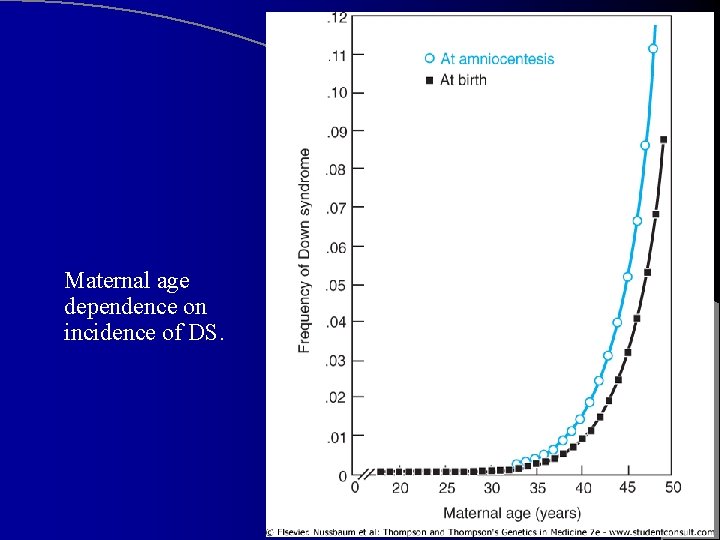
Down Syndrome (Trisomy 21) l Most common genetic cause of moderate MR l ~ 1 child in 800 is born with DS. Incidence is higher in mothers >35 yrs. l Two noteworthy features of its pop. distribution: – Increased maternal age – Peculiar distribution within families: concordance in monozygotic twins but almost complete discordance in dizygotic twins and other family members

Maternal age dependence on incidence of DS.

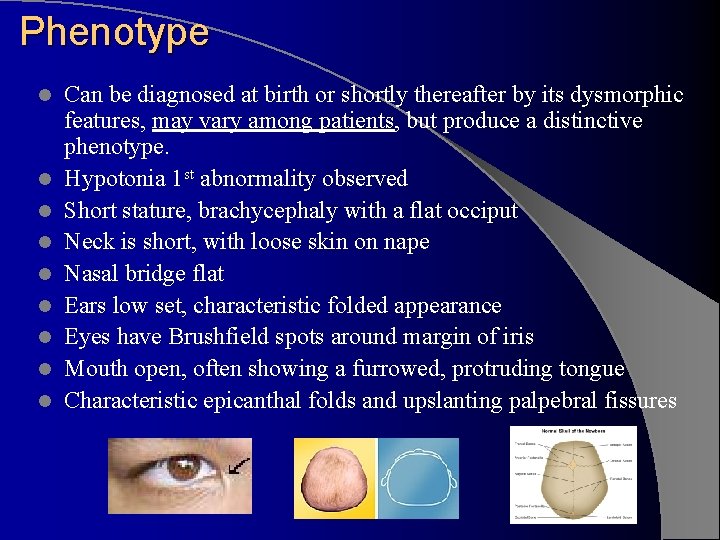
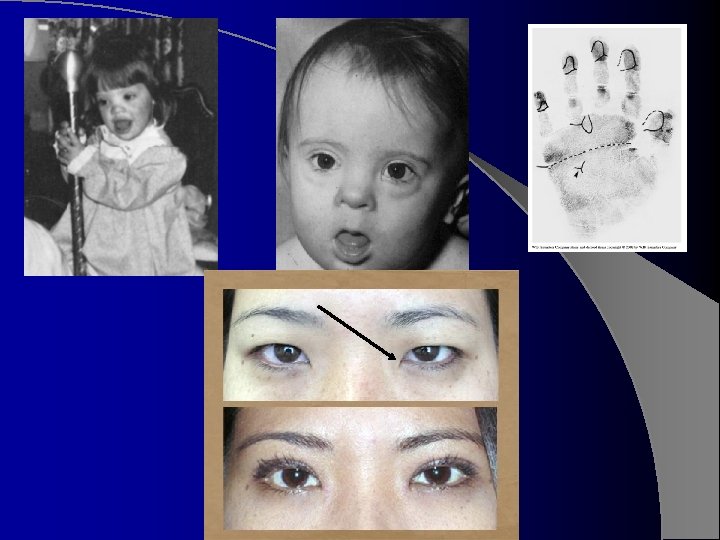
Phenotype l l l l l Can be diagnosed at birth or shortly thereafter by its dysmorphic features, may vary among patients, but produce a distinctive phenotype. Hypotonia 1 st abnormality observed Short stature, brachycephaly with a flat occiput Neck is short, with loose skin on nape Nasal bridge flat Ears low set, characteristic folded appearance Eyes have Brushfield spots around margin of iris Mouth open, often showing a furrowed, protruding tongue Characteristic epicanthal folds and upslanting palpebral fissures


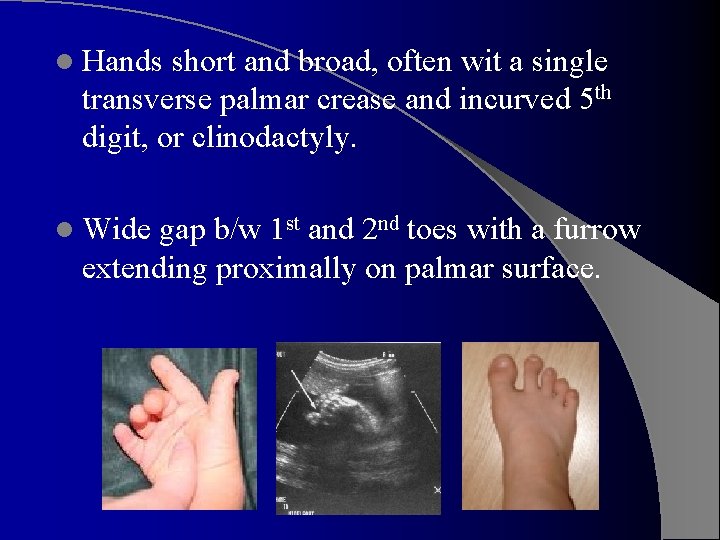
l Hands short and broad, often wit a single transverse palmar crease and incurved 5 th digit, or clinodactyly. l Wide gap b/w 1 st and 2 nd toes with a furrow extending proximally on palmar surface.

Major concern is MR. developmental delay is usually obvious by end of 1 st year. IQ is usually 30 to 60. l Congenital heart disease in at least 1/3 of DS infants (higher in abortuses) l There is a high degree of variability in phenotype; specific abnormalities detected in almost all, others seen only in subset of cases. l Birth defects reflect effects of overexpression of gene(s) during early development e. g. , a significant proportion of chr. 21 genes are expressed at higher levels in DS brain and heart samples as compared to euploid individuals. l

Prenatal and Postnatal Survival At all maternal ages, there is some loss b/w 11 th and 16 th wks, and an additional loss later in pregnancy. l Probably ~ 20 -25% of trisomy 21 survive to birth. l Among DS coceptuses, l – those least likely to survive are those with congenital heart disease – ~ ¼ of liveborns with heart defects die before 1 st birthday There is 15 -fold increase in risk of leukemia among DS who survive neonatal period l Premature dementia, associated with neuropathological findings characteristic of Alzheimer disease affects DS patients several decades earlier than in general pop. l

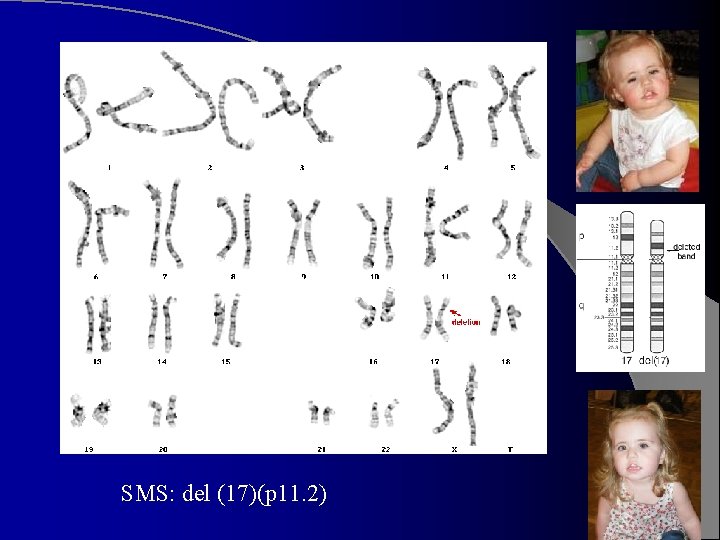
The chromosomes in DS Trisomy 21 Karyotyoing is necessary for confirmation and basis for genetic counseling. It is essential for determining recurrence risk. l In ~ 95%, 47, XX/XY, +21, resulting from meiotic non-disjunction. l Risk of having a child with trisomy 21 increases with maternal age, esp. after 30. l Meiotic error usually maternal (~90% of cases), predominantly meiosis-I. ~ 10% paternal, usually meiosis-II. l

Robertsonian Translocation ~ 4% of DS, 46 chr’s one of which a Rob translocation b/w 21 q and q arm of another acrocentric (usually 14 or 22). E. g. , 46, xx, rob(14; 21)(q 10; q 10), +21. Such a chr is a. k. a der(14; 21). In effect patient is trisomic for 21 q l No relation to maternal age l Has a relatively high recurrence risk in families esp. when mother is the carrier l Karyotyping for parents and other relatives is essential for accurate counseling l

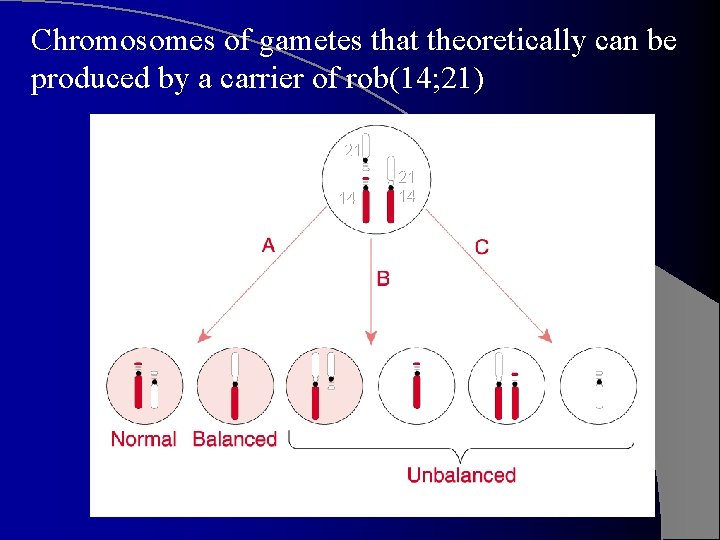
Carrier has 45 chr’s. l There are 6 possible types of gametes, 3 of which do not lead to viable offspring. l Of the 3 viable: one is normal, one is balanced and one is unbalanced. Theoretically produced in equal numbers. l Studies have shown that unbalanced chr. complement appears in ~ 10 -15% of the progeny of a carrier mother (only a few % when father is the carrier). l

Chromosomes of gametes that theoretically can be produced by a carrier of rob(14; 21) 21 14

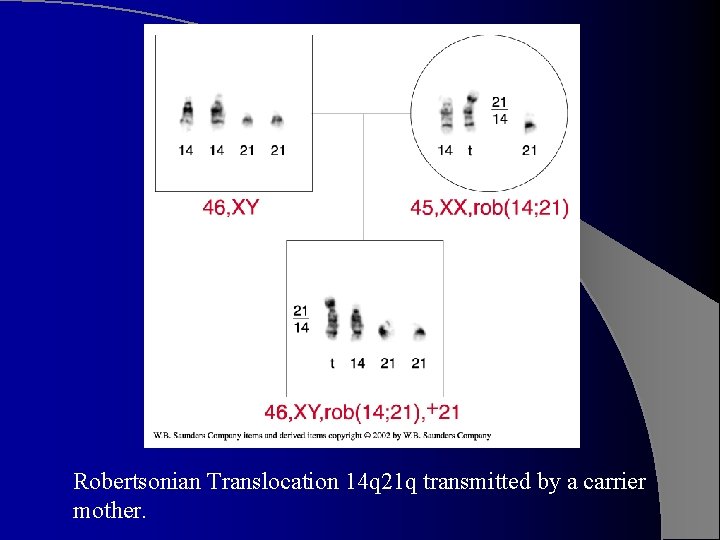
Robertsonian Translocation 14 q 21 q transmitted by a carrier mother.

21 q translocation Seen in a few % of DS Thought to originate as an isochromosome Many cases arise post-zygotically, hence recurrence risk is low. l Still, important to evaluate if a parent is a carrier (or mosaic). What type of gametes would a 21 q carrier produce? What about potential progeny? l Mosaic carriers are at increased risk of recurrence. Prenatal diagnosis should be considered. l l l

Mosaic DS l~ 2% of DS, usually with normal/trisomy 21 karyotype. l Phenotype may be milder, with wide variability among mosaic patients. l Mildly affected are less likely to be karyotyped Partial Trisomy 21 l Part of 21 q is present in triplicate, very rare. l DS with no cytogenet. visible chr. abnormality is even more rarely identified.

Etiology of Trisomy 21 Something about maternal meiosis-I is the underlying cause in high % of trisomy 21. l “older egg” model: the older the egg, the greater the chance that chr’s will fail to disjoin correctly. l Older eggs may be less able to overcome a susceptibility to non-disjunction established by recombination (recall recombination pattern and non-disjunction) machinery. l Etiological events may have taken place many years ago, when mother’s primary oocytes were still in prophase of meiosis-I. l

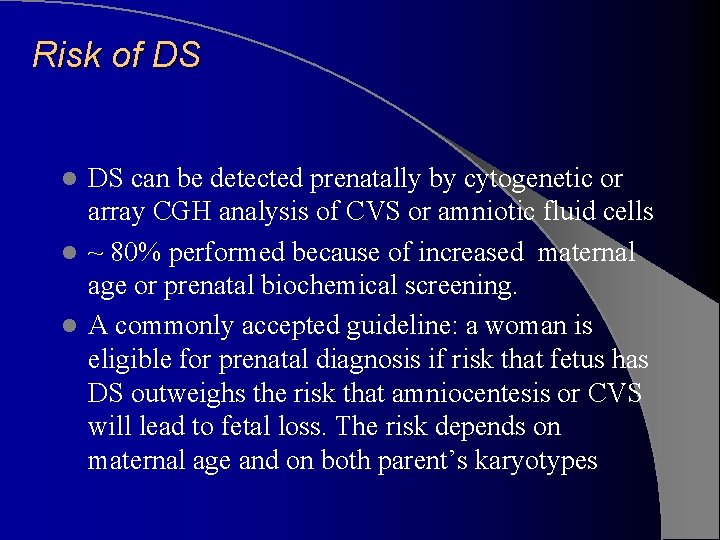
Risk of DS DS can be detected prenatally by cytogenetic or array CGH analysis of CVS or amniotic fluid cells l ~ 80% performed because of increased maternal age or prenatal biochemical screening. l A commonly accepted guideline: a woman is eligible for prenatal diagnosis if risk that fetus has DS outweighs the risk that amniocentesis or CVS will lead to fetal loss. The risk depends on maternal age and on both parent’s karyotypes l

l l l Pop. incidence of DS in live births is ~ 1 in 800. At ~ age 30, risk begins to rise sharply, reaching 1 in 25 births in oldest maternal age group. Although younger mothers have lower risk, their birth rate is much higher, and therefore > ½ of mothers of all DS babies are <35. Risk of DS due to translocation or partial trisomy is unrelated to maternal age Paternal age seems to have no influence on risk

Recurrence risk l l l Recurrence risk trisomy 21 (or some other autosomal trisomy), after one such child has been born in a family is ~ 1% overall. The risk is ~ 1. 4% for mothers <30 Reason for increased risk for younger women in not known (one possibility is unrecognized germline mosaicism in one parent). A history of trisomy 21 elsewhere in the family does not appear to increase risk of having a DS child. Recurrence risk for DS due to translocation is much higher

Trisomy 18 – Edwards Syndrome l Features include: MR and failure to thrive Often, severe malformation of heart Hypertonia Head has prominent occiput, and jaw recedes Ears low-set and malformed Sternum short Clenched fist, with 2 nd and 5 th digits overlapping 3 rd and 4 th. – “Rocker bottom” appearance of feet – Single creases on palms and arch patterns on most or all digits. – – – –

Trisomy 18 – Edwards syndrome 1/7500 live births Karyotype: 47, XX (or XY), +18 Prenatal detection: strong association with abnormal anatomy – choroid plexus cysts, CNS malformation, CHD (ventricular septal & outflow tract defects) abnormalities

l l l l l Incidence in liveborns is 1 in 7500 births Incidence at conception is much higher, but ~ 95% are aborted spont. Survival for > a few months is rare At least 60% of patients female (preferential survival? ) Increased maternal age is a factor. Trisomy 18 phenotype, like trisomy 21, can result from a variety of rare karyotypes other than complete trisomy. Karyotyping of affected infants/fetuses is essential for counseling In ~ 20%, translocation involving all or most chr. 18 (either de novo or inherited from a balanced carrier) Trisomy 18 may be mosaic, with variable but usually milder expression

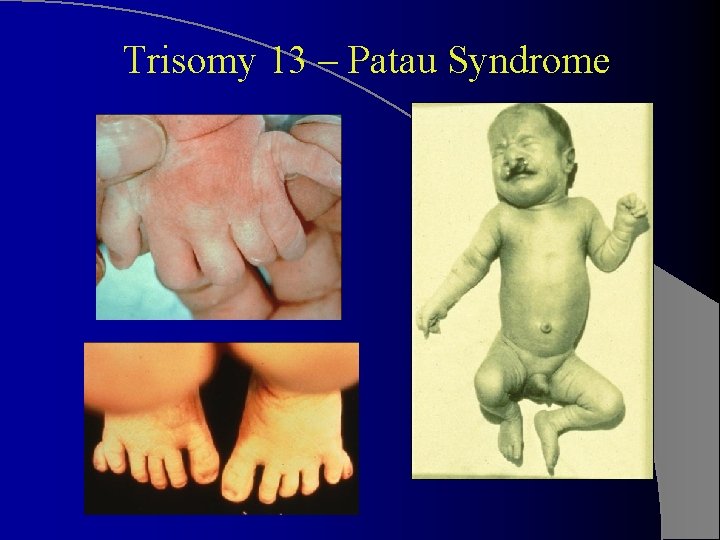
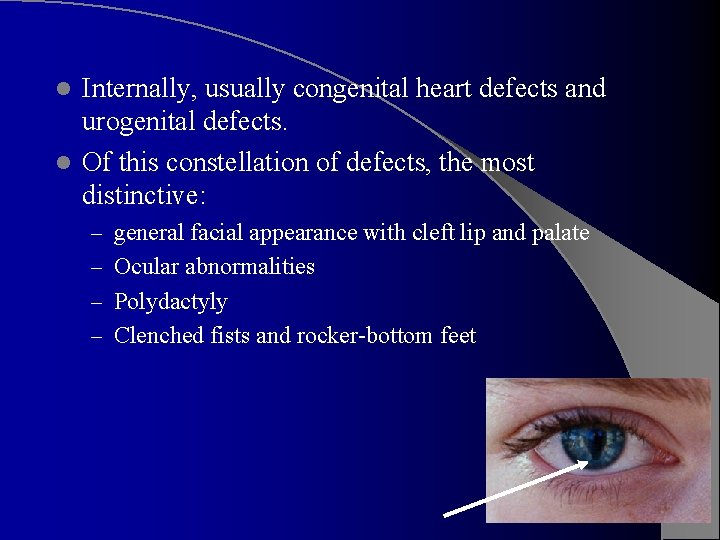
Trisomy 13 – Patau Syndrome Growth retardation and severe MR, accompanied by severe CNS malformations. l Forehead is sloping; microcephaly and wideopen sutures; may be micophthalmia, iris coloboma, or even absence of the eye l Ears malformed l Cleft lip and cleft palate are often present l Hands and feet may show postaxial polydactyly, and hands clench as in trisomy 18 l Feet as in trisomy 18, “rocker bottom” appearance l Palms often have simian crease l

Trisomy 13 – Patau Syndrome

Internally, usually congenital heart defects and urogenital defects. l Of this constellation of defects, the most distinctive: l – general facial appearance with cleft lip and palate – Ocular abnormalities – Polydactyly – Clenched fists and rocker-bottom feet

l l l Incidence is ~ 1 in 15, 000 -25, 000 births About ½ die within 1 st month Associated with increased maternal age, and extra chr. usually from nondisjunction in maternal meiosis-I. Karyotyping is indicated to confirm diagnosis ~ 20% caused by unbalanced translocation Recurrence risk is low; even when one parent is a carrier of translocation, empirical risk that a subsequent child will have the syndrome is <2%.

Autosomal Deletion Syndromes l Overall, cytogenetically visible autosomal deletions occur with an estimated incidence of 1/7000 live births. Cri du Chat Syndrome l Terminal or interstitial deletion of part of 5 p. l Name was given because crying infants sound like a mewing cat.

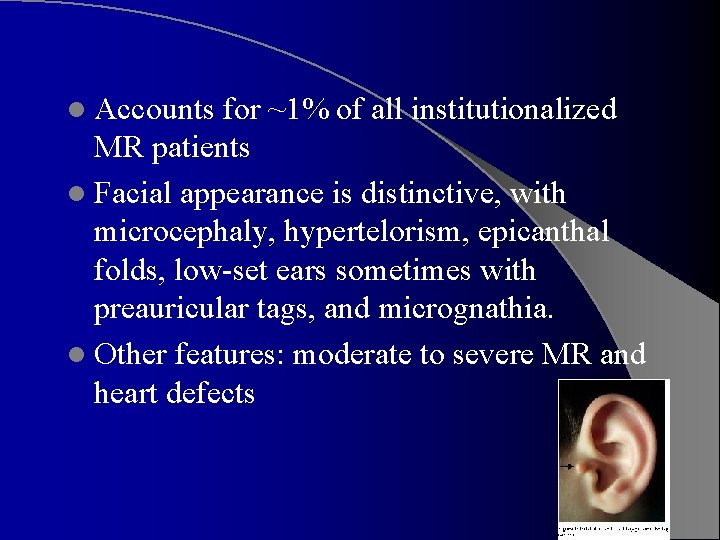
l Accounts for ~1% of all institutionalized MR patients l Facial appearance is distinctive, with microcephaly, hypertelorism, epicanthal folds, low-set ears sometimes with preauricular tags, and micrognathia. l Other features: moderate to severe MR and heart defects

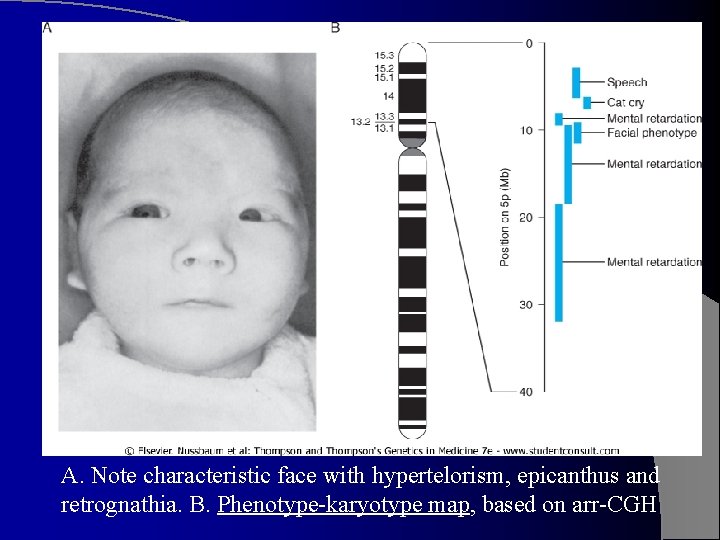
A. Note characteristic face with hypertelorism, epicanthus and retrognathia. B. Phenotype-karyotype map, based on arr-CGH

Most cases are sporadic; 10 -15% are offspring of translocation carriers. l Breakpoints and extent of deleted segment of 5 p vary in patients, critical region missing in all is 5 p 15 l Many of the phenotypes due to haploinsufficiency for gene(s) within 5 p 15. 2, and distinctive cat cry from deletion of gene(s) within a small region in band 5 p 15. 3. l Degree of MR usually correlates with size of deletion. l

Genomic Disorders: Microdeletion and Duplication Syndromes Many dysmorphic syndromes associated with small (sometimes cytogenet. visible) deletions that lead to a form of genetic imbalance referred to as segmental aneusomy. l These deletions produce clinically recognizable syndromes. Can be detected by high-resolution banding, FISH, arr-CGH l The term contiguous gene syndrome has been applied to many of them. i. e. , haploinsuf. for multiple contiguous genes within deleted region l For other disorders, phenotype is apparently due to deletion of a single gene, despite association of a chr. deletion with the condition. l

l For each syndrome extent of del in different patients is similar, suggesting existence of deletion-prone sequences. l Fine mapping has shown that breakpoints localize to low-copy repeated sequences and that aberrant recombination b/w nearby copies of the repeats causes deletions (which span several 100 -1000 kb). l This general sequence-dependent mechanism has been implicated in several contiguous gene rearrangement syndromes, which have therefore been termed genomic disorders.

Several deletions/duplications mediated by unequal recombination have been documented within proximal 17 p. l E. g. , a cytogenet. visible segment of 17 p 11. 2 of ~ 4 Mb is deleted de novo in ~ 70 -80% of patients with Smith-Magenis syndrome (SMS), a usually sporadic condition characterized by multiple congenital anomalies and MR. l The major features of SMS include mild to moderate mental retardation, delayed speech and language skills, distinctive facial features, sleep disturbances, and behavioral problems. l

l Unequal recombination b/w large blocks of flanking repeated sequences (nearly 99% identical) results in SMS deletion, del(17)(p 11. 2 -p 11. 2), as well as the reciprocal duplication dup(17)(p 11. 2), seen in patients with a milder, neurobehavioral phenotype

SMS: del (17)(p 11. 2)

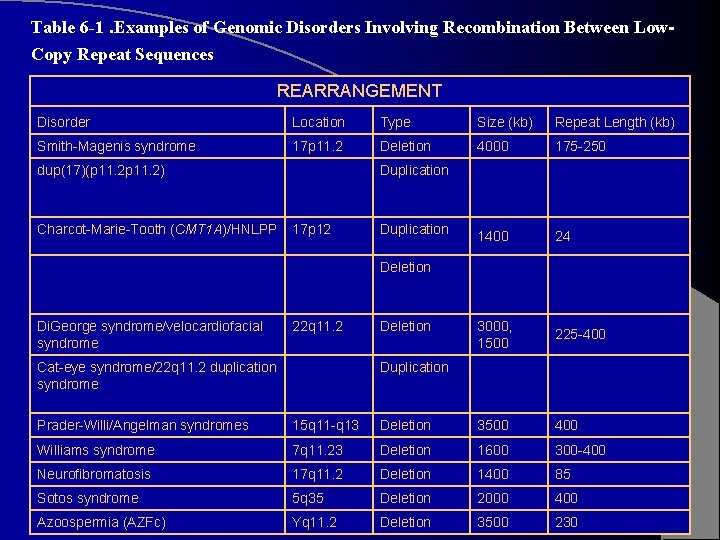
Table 6 -1. Examples of Genomic Disorders Involving Recombination Between Low. Copy Repeat Sequences REARRANGEMENT Disorder Location Type Size (kb) Repeat Length (kb) Smith-Magenis syndrome 17 p 11. 2 Deletion 4000 175 -250 dup(17)(p 11. 2) Duplication Charcot-Marie-Tooth (CMT 1 A)/HNLPP 17 p 12 Duplication 1400 24 3000, 1500 225 -400 Deletion Di. George syndrome/velocardiofacial syndrome 22 q 11. 2 Deletion Cat-eye syndrome/22 q 11. 2 duplication syndrome Duplication Prader-Willi/Angelman syndromes 15 q 11 -q 13 Deletion 3500 400 Williams syndrome 7 q 11. 23 Deletion 1600 300 -400 Neurofibromatosis 17 q 11. 2 Deletion 1400 85 Sotos syndrome 5 q 35 Deletion 2000 400 Azoospermia (AZFc) Yq 11. 2 Deletion 3500 230

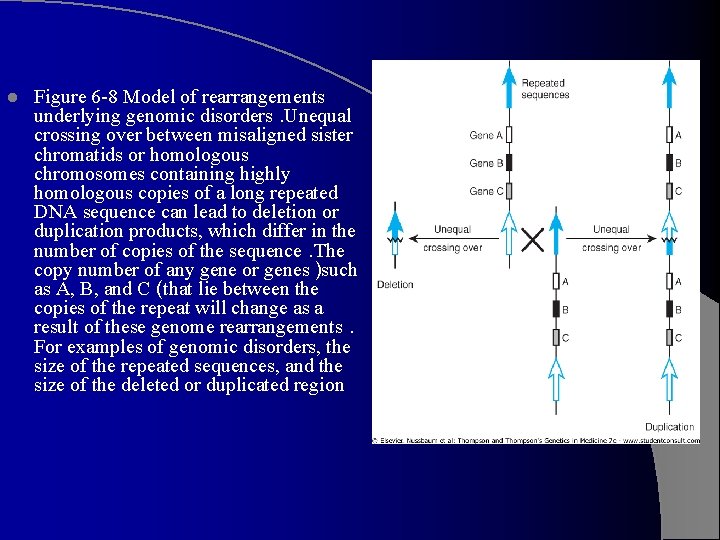
l Figure 6 -8 Model of rearrangements underlying genomic disorders. Unequal crossing over between misaligned sister chromatids or homologous chromosomes containing highly homologous copies of a long repeated DNA sequence can lead to deletion or duplication products, which differ in the number of copies of the sequence. The copy number of any gene or genes )such as A, B, and C (that lie between the copies of the repeat will change as a result of these genome rearrangements. For examples of genomic disorders, the size of the repeated sequences, and the size of the deleted or duplicated region

Di. George syndrome A particularly common microdeletion that is frequently evaluated in clinical cytogenetics laboratories involves chromosome 22 q 11. 2 and is associated with diagnoses of Di. George syndrome, velocardiofacial syndrome, or conotruncal anomaly face syndrome. l All three clinical syndromes are autosomal dominant conditions with variable expressivity, caused by a deletion within 22 q 11. 2, spanning about 3 Mb. This microdeletion, also mediated by homologous recombination between lowcopy repeated sequences, is one of the most common cytogenetic deletions associated with an important clinical phenotype and is detected in 1 in 2000 to 4000 live births (Fig. 6 -9). l Patients show characteristic craniofacial anomalies, mental retardation, and heart defects. The deletion in the 22 q 11. 2 deletion syndromes is thought to play a role in as many as 5% of all congenital heart defects and is a particularly frequent cause of certain defects. l

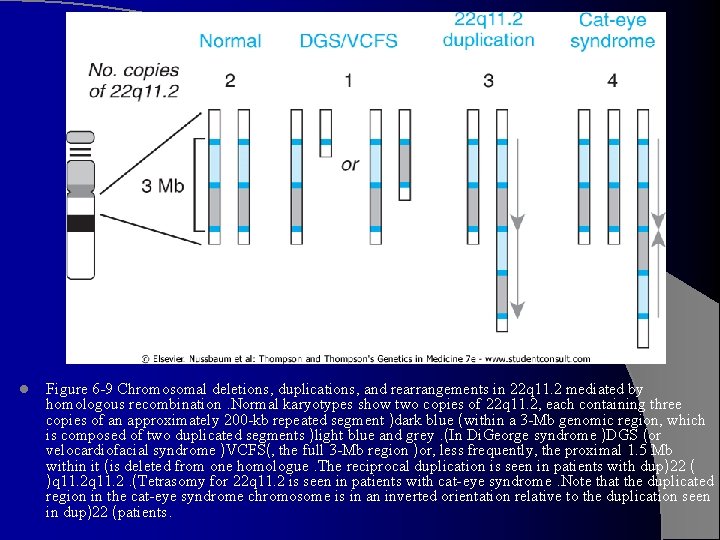
l Figure 6 -9 Chromosomal deletions, duplications, and rearrangements in 22 q 11. 2 mediated by homologous recombination. Normal karyotypes show two copies of 22 q 11. 2, each containing three copies of an approximately 200 -kb repeated segment )dark blue (within a 3 -Mb genomic region, which is composed of two duplicated segments )light blue and grey. (In Di. George syndrome )DGS (or velocardiofacial syndrome )VCFS(, the full 3 -Mb region )or, less frequently, the proximal 1. 5 Mb within it (is deleted from one homologue. The reciprocal duplication is seen in patients with dup)22 ( )q 11. 2. (Tetrasomy for 22 q 11. 2 is seen in patients with cat-eye syndrome. Note that the duplicated region in the cat-eye syndrome chromosome is in an inverted orientation relative to the duplication seen in dup)22 (patients.

l The typical deletion removes approximately 30 genes, although a related, smaller deletion is seen in approximately 10% of cases. l Haploinsufficiency for at least one of these genes, TBX 1, which encodes a transcription factor involved in development of the pharyngeal system, has been implicated in the phenotype; it is contained within the deleted region and is mutated in patients with a similar phenotype but without the chromosomal deletion.

In contrast to the relatively common deletion of 22 q 11. 2, the reciprocal duplication of 22 q 11. 2 is much rarer and leads to a series of dysmorphic malformations and birth defects called the 22 q 11. 2 duplication syndrome. l Diagnosis of this duplication generally requires analysis by FISH on interphase cells or array CGH. l Some patients have a quadruple complement of this segment of chromosome 22 and are said to have cateye syndrome, which is characterized clinically by ocular coloboma, congenital heart defects, craniofacial anomalies, and moderate mental retardation. l The karyotype in cat-eye syndrome is 47, XX or XY, +inv dup(22)(pter→q 11. 2). l

The constellation of different disorders associated with varying dosage of genes in this segment of chromosome 22) see Fig. 6 -9) reflects two major principles in clinical cytogenetics. l First, with few exceptions, altered gene dosage for any extensive chromosomal or genomic region is likely to result in a clinical abnormality, the phenotype of which will, in principle, depend on haploinsufficiency for or overexpression of one or more genes encoded within the region. l Second, even patients carrying what appears to be the same chromosomal deletion or duplication can present with a range of variable phenotypes. Although the precise basis for this variability is unknown, it could be due to non-genetic causes or to differences in the genome sequence among unrelated individuals. l
 Autosomal recessive and dominant
Autosomal recessive and dominant Alleles are
Alleles are Naacls accredited cytogenetics education program
Naacls accredited cytogenetics education program Cytogenetics
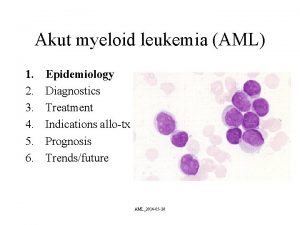
Cytogenetics Aml survival by age
Aml survival by age Autosomes and sex chromosomes
Autosomes and sex chromosomes Autosomes
Autosomes Two copies of each autosomal gene affect
Two copies of each autosomal gene affect Autosomes
Autosomes How many autosomes are there in a human sperm? *
How many autosomes are there in a human sperm? * Autosomes in karyotypes
Autosomes in karyotypes Whats a autosome
Whats a autosome Autosomes vs sex chromosomes
Autosomes vs sex chromosomes Autosomes
Autosomes Gamete and zygote
Gamete and zygote Is down syndrome autosomal or sexlinked
Is down syndrome autosomal or sexlinked Tautan dan pindah silang
Tautan dan pindah silang Jacobs syndrome
Jacobs syndrome Mendelian dihybrid test cross
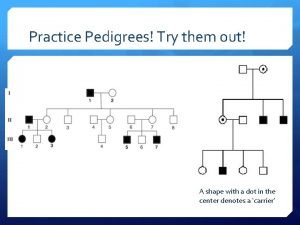
Mendelian dihybrid test cross Pedigree symbols
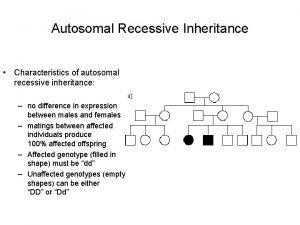
Pedigree symbols Autosomal recessive
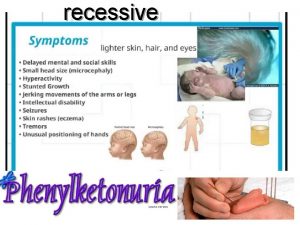
Autosomal recessive Pku recessive or dominant
Pku recessive or dominant Autosomal dominant
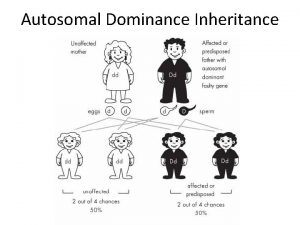
Autosomal dominant Autosomal dominant punnett square
Autosomal dominant punnett square Generations in a pedigree
Generations in a pedigree Is klinefelter syndrome autosomal or sexlinked
Is klinefelter syndrome autosomal or sexlinked Trisomy 13 pictures
Trisomy 13 pictures Immune system structure
Immune system structure Example of sex linked pedigree
Example of sex linked pedigree Is color blindness autosomal or sexlinked
Is color blindness autosomal or sexlinked Two copies of each autosomal gene affect phenotype
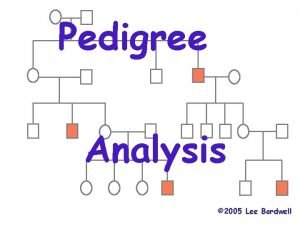
Two copies of each autosomal gene affect phenotype Pedegree
Pedegree Hereditary spherocytosis autosomal
Hereditary spherocytosis autosomal Flurorescein
Flurorescein Is methemoglobinemia autosomal or sexlinked
Is methemoglobinemia autosomal or sexlinked Autosomal recessive cystic fibrosis
Autosomal recessive cystic fibrosis Recessive trait
Recessive trait Sexlinked vs autosomal
Sexlinked vs autosomal What are sexlinked traits
What are sexlinked traits What are sexlinked traits
What are sexlinked traits Chorionic villus sampling
Chorionic villus sampling Hair on end appearance
Hair on end appearance What is karyotyping
What is karyotyping Leigh's disease
Leigh's disease