Chloroplast Genome Evolution Level 3 Molecular Evolution and



- Slides: 13

Chloroplast Genome Evolution Level 3 Molecular Evolution and Bioinformatics Jim Provan

References Douglas SE (1998) “Plastid evolution: origins, diversity, trends” Current Opinion in Genetics and Development 8: 655 -661 Sugiura M (1995) “The chloroplast genome” Essays in Biochemistry 30: 49 -57 Gray MW (1993) “Origin and evolution of organelle genomes” Current Opinion in Genetics and Development 3: 884 -890

The chloroplast Carries out photosynthesis Contains own genome Believed to be of endosymbiotic origin Phylogenetically related to cyanobacteria

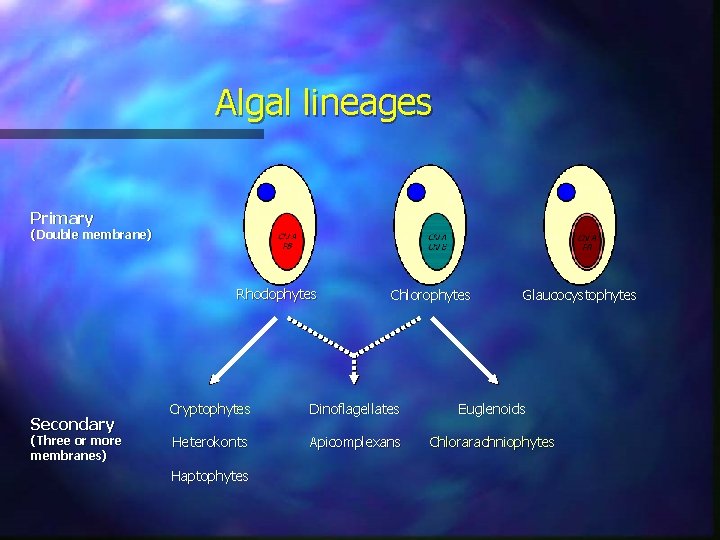
Algal lineages Primary (Double membrane) Rhodophytes Secondary (Three or more membranes) Chlorophytes Glaucocystophytes Cryptophytes Dinoflagellates Euglenoids Heterokonts Apicomplexans Chlorarachniophytes Haptophytes

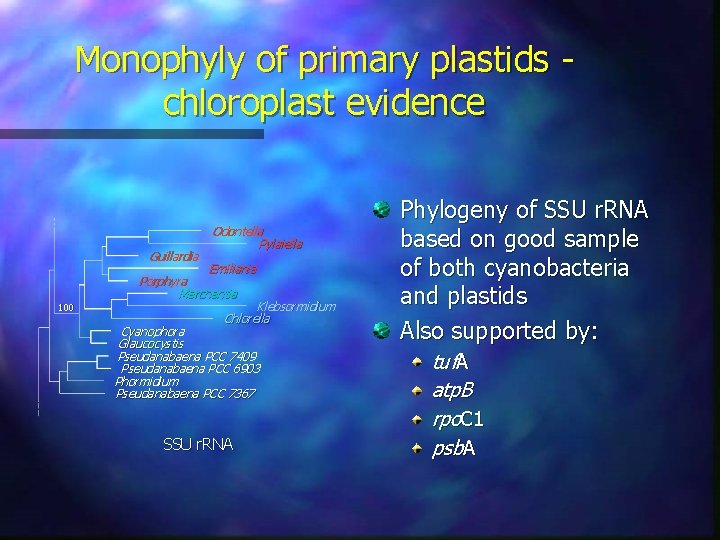
Monophyly of primary plastids chloroplast evidence Guillardia 100 Odontella Pylaiella Emiliania Porphyra Marchantia Klebsormidium Chlorella Cyanophora Glaucocystis Pseudanabaena PCC 7409 Pseudanabaena PCC 6903 Phormidium Pseudanabaena PCC 7367 SSU r. RNA Phylogeny of SSU r. RNA based on good sample of both cyanobacteria and plastids Also supported by: tuf. A atp. B rpo. C 1 psb. A

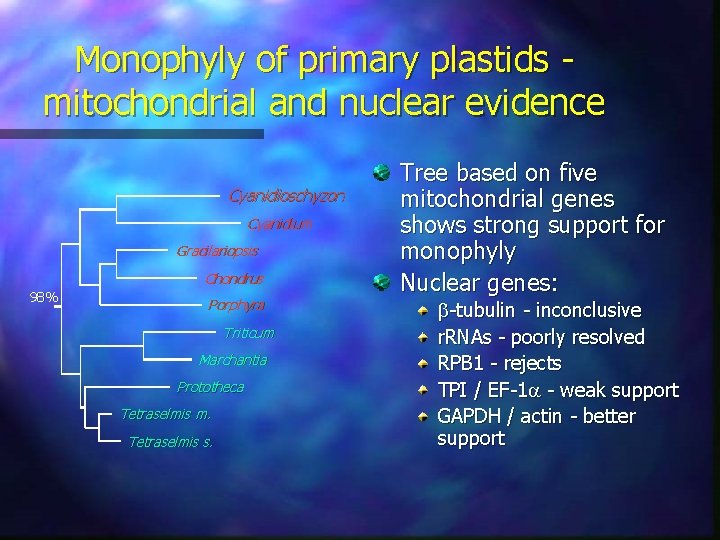
Monophyly of primary plastids mitochondrial and nuclear evidence Cyanidioschyzon Cyanidium Gracilariopsis Chondrus 98% Porphyra Triticum Marchantia Prototheca Tetraselmis m. Tetraselmis s. Tree based on five mitochondrial genes shows strong support for monophyly Nuclear genes: -tubulin - inconclusive r. RNAs - poorly resolved RPB 1 - rejects TPI / EF-1 - weak support GAPDH / actin - better support

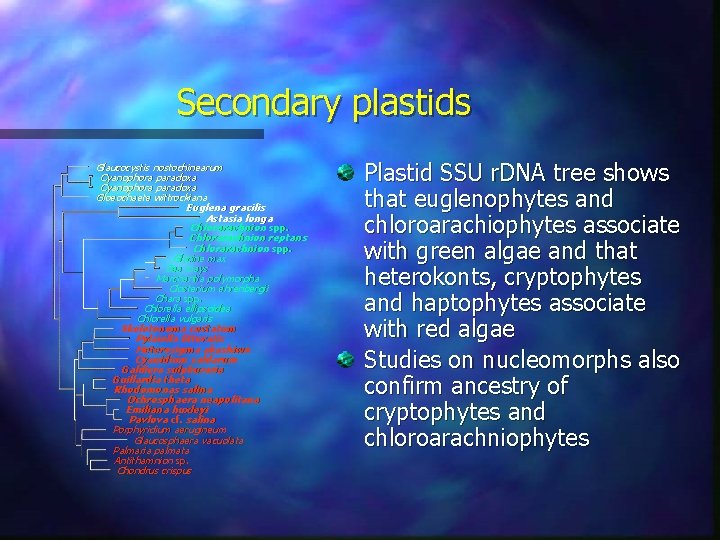
Secondary plastids Glaucocystis nostochinearum Cyanophora paradoxa Gloeochaete wittrockiana Euglena gracilis Astasia longa Chlorarachnion spp. Chlorarachnion reptans Chlorarachnion spp. Glycine max Zea mays Marchantia polymorpha Closterium ehrenbergii Chara spp. Chlorella ellipsoidea Chlorella vulgaris Skeletonema costatum Pylaiella littoratis Heterosigma akashiwo Cyanidium caldarum Galdiera sulphuraria Guillardia theta Rhodomonas salina Ochrosphaera neapolitana Emiliana huxleyi Pavlova cf. salina Porphyridium aerugineum Glaucosphaera vacuolata Palmaria palmata Antithamnion sp. Chondrus crispus Plastid SSU r. DNA tree shows that euglenophytes and chloroarachiophytes associate with green algae and that heterokonts, cryptophytes and haptophytes associate with red algae Studies on nucleomorphs also confirm ancestry of cryptophytes and chloroarachniophytes

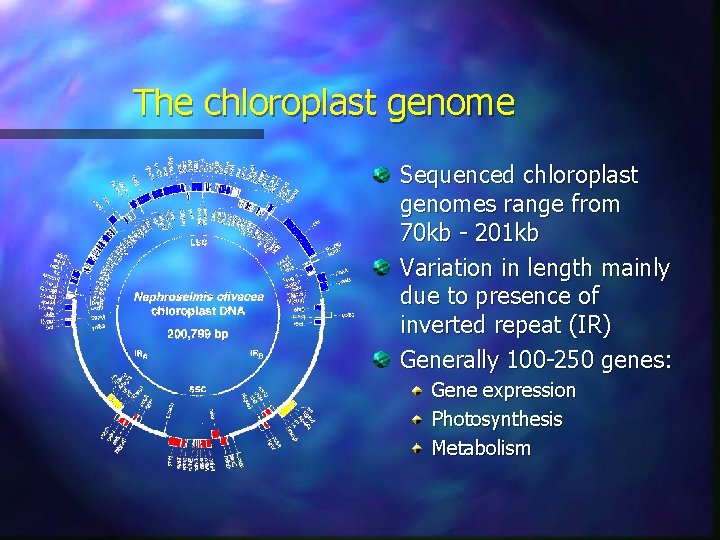
The chloroplast genome Sequenced chloroplast genomes range from 70 kb - 201 kb Variation in length mainly due to presence of inverted repeat (IR) Generally 100 -250 genes: Gene expression Photosynthesis Metabolism

The inverted repeat (IR) Pinus Ranges from 5 bk to 76 kb in length IR contains r. RNA genes plus others: None in brown algae (5 kb) 10 in tobacco (25 kb) 40 in geranium (76 kb) Nicotiana Present in: Porphyra Land plants (exc. legumes) Chlorophytes Chromophytes Partial in conifers

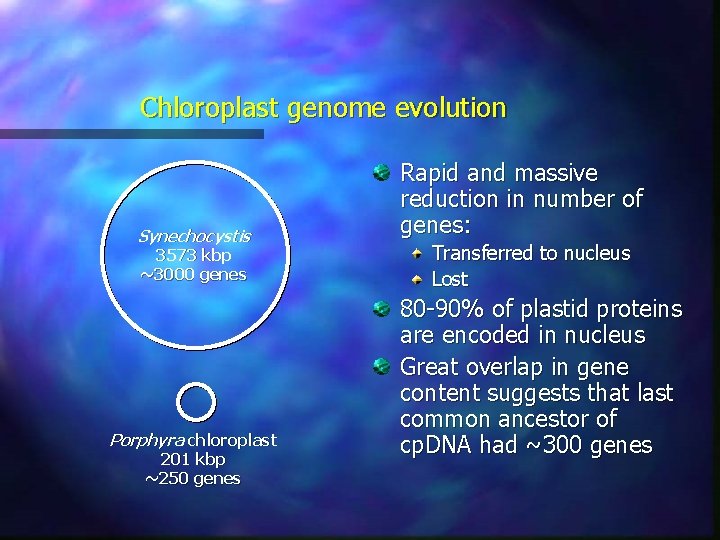
Chloroplast genome evolution Synechocystis 3573 kbp ~3000 genes Porphyra chloroplast 201 kbp ~250 genes Rapid and massive reduction in number of genes: Transferred to nucleus Lost 80 -90% of plastid proteins are encoded in nucleus Great overlap in gene content suggests that last common ancestor of cp. DNA had ~300 genes

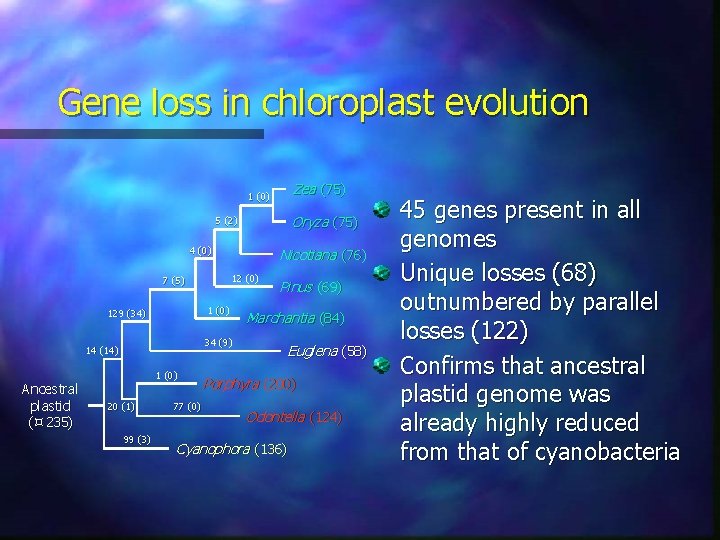
Gene loss in chloroplast evolution Zea (75) 1 (0) Oryza (75) 5 (2) 4 (0) 12 (0) 7 (5) 1 (0) 129 (34) 34 (9) 14 (14) Ancestral plastid ( 235) Nicotiana (76) 1 (0) 20 (1) 99 (3) 77 (0) Pinus (69) Marchantia (84) Euglena (58) Porphyra (200) Odontella (124) Cyanophora (136) 45 genes present in all genomes Unique losses (68) outnumbered by parallel losses (122) Confirms that ancestral plastid genome was already highly reduced from that of cyanobacteria

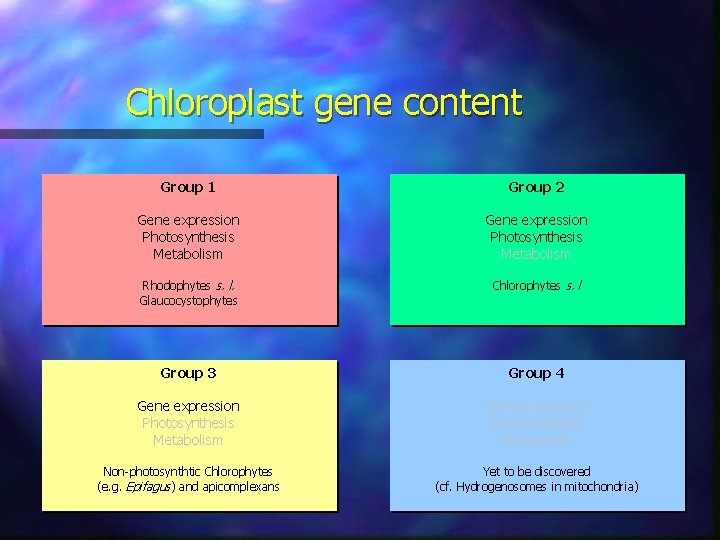
Chloroplast gene content Group 1 Group 2 Gene expression Photosynthesis Metabolism Rhodophytes s. l. Glaucocystophytes Chlorophytes s. l Group 3 Group 4 Gene expression Photosynthesis Metabolism Non-photosynthtic Chlorophytes (e. g. Epifagus) and apicomplexans Yet to be discovered (cf. Hydrogenosomes in mitochondria)

Chloroplast genes (excluding ycf) Land Plants Epifagus Euglena Others Porphyra 101 -107 40 82 113 -166 181 4 30 -32 2 -21 5 -6 4 17 15 2 3 27 21 4 3 28 -36 21 -44 6 -9 3 35 47 18 29 -30 11 - 26 - 31 -48 -/10 53 - 1 -5 2 1 7 -14 25 18 -21 6 149+ -/1/3 - Photosyn. Total Gene expression r. RNA t. RNA r-protein Other Photosynthesis Ru. Bis. Co/thylakoid ndh Metabolism/misc. Introns Algae