CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE Highthroughput comparison


- Slides: 20

CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE High-throughput comparison of Duggable Protein-Ligand Binding sites Didier Rognan Institut Gilbert Lasutriat Bioinformatics of the Drug National Center for Scientific Research (CNRS) F-67400 Illkirch, France didier. rognan@pharma. u-strasbg. fr

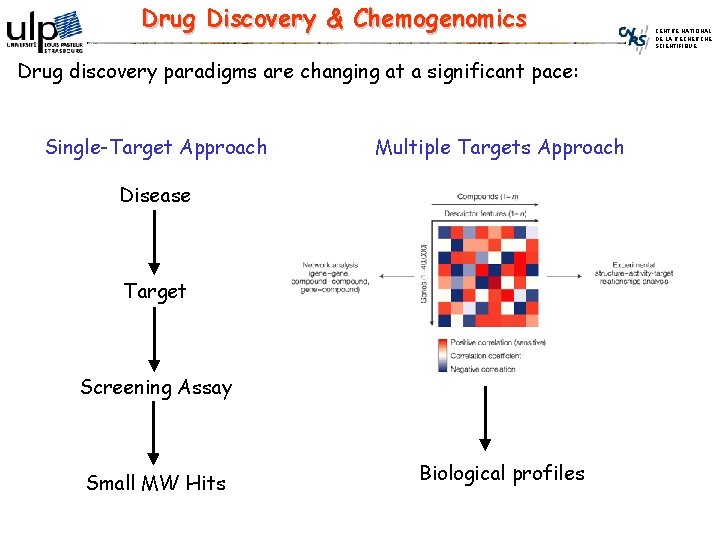
Drug Discovery & Chemogenomics Drug discovery paradigms are changing at a significant pace: Single-Target Approach Multiple Targets Approach Disease Target Screening Assay Small MW Hits Biological profiles CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE

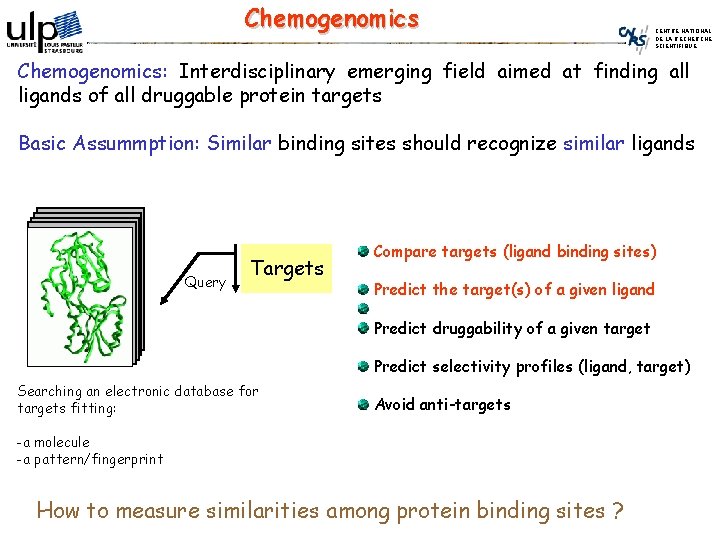
Chemogenomics CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE Chemogenomics: Interdisciplinary emerging field aimed at finding all ligands of all druggable protein targets Basic Assummption: Similar binding sites should recognize similar ligands Query Targets Compare targets (ligand binding sites) Predict the target(s) of a given ligand Predict druggability of a given target Predict selectivity profiles (ligand, target) Searching an electronic database for targets fitting: Avoid anti-targets -a molecule -a pattern/fingerprint How to measure similarities among protein binding sites ?

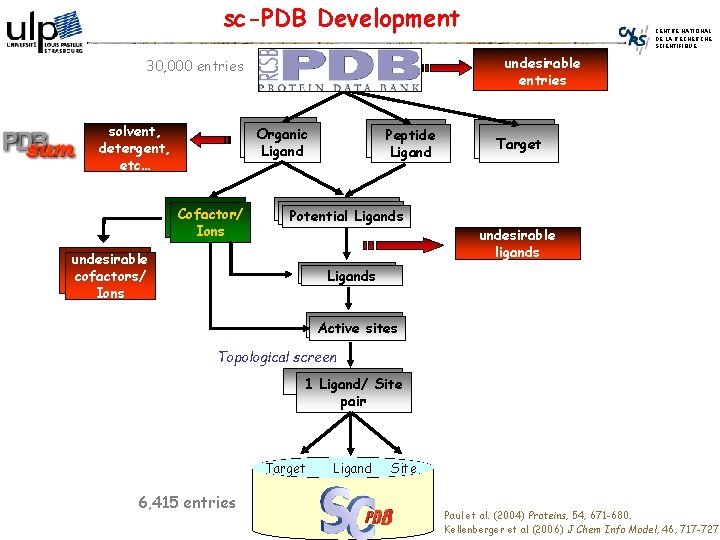
sc-PDB Development undesirable entries 30, 000 entries solvent, detergent, etc… Organic Ligand Cofactor/ Ions CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE Peptide Ligand Potential Ligands undesirable cofactors/ Ions Target undesirable ligands Ligands Active sites Topological screen 1 Ligand/ Site pair Target 6, 415 entries Ligand Site Paul et al. (2004) Proteins, 54, 671 -680. Kellenberger et al (2006) J Chem Info Model, 46, 717 -727.

sc. PDB: Comparison of binding sites CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE Ligand descriptors cannot be used to describe protein binding sites and measure their similarities How to measure the distance between two binding sites ? Sequenced-based approaches (GASH, Dali. Lite, CE) Aligning fragment pairs in overlapping windows Computing NERs (number of equivalent residues) What about active sites (discontinuous sequences) ? How to detect local similarity in absence of global sequence identity ? Structure-based approaches (Cavbase, Site. Engine, Sumo, Sitesbase) Surface-based physchem properties (pseudo-centers, triplets) maximal common subgraph matching Atom coordinates sensitivity (homology models) ? Normalized score (0 1) ? Binding site druggability ?

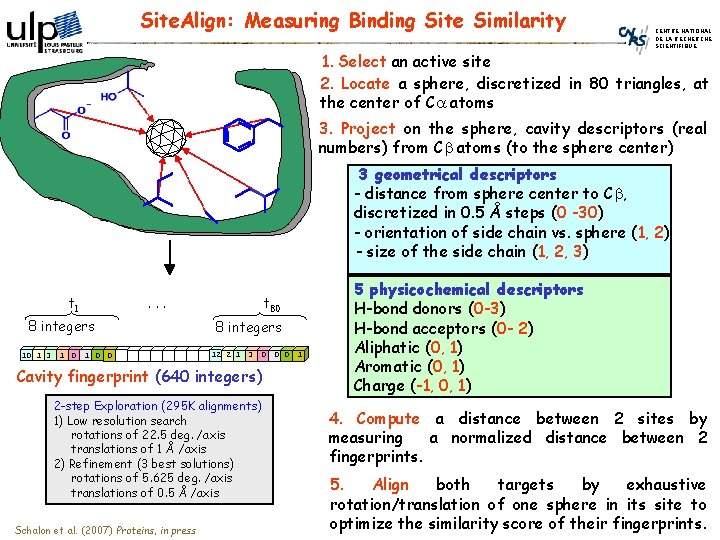
Site. Align: Measuring Binding Site Similarity CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE 1. Select an active site 2. Locate a sphere, discretized in 80 triangles, at the center of Ca atoms 3. Project on the sphere, cavity descriptors (real numbers) from Cb atoms (to the sphere center) 3 geometrical descriptors - distance from sphere center to Cb, discretized in 0. 5 Å steps (0 -30) - orientation of side chain vs. sphere (1, 2) - size of the side chain (1, 2, 3) t 1 . . . 8 integers 10 1 3 1 0 0 t 80 8 integers 12 2 1 3 0 0 0 1 Cavity fingerprint (640 integers) 2 -step Exploration (295 K alignments) 1) Low resolution search rotations of 22. 5 deg. /axis translations of 1 Å /axis 2) Refinement (3 best solutions) rotations of 5. 625 deg. /axis translations of 0. 5 Å /axis Schalon et al. (2007) Proteins, in press 5 physicochemical descriptors H-bond donors (0 -3) H-bond acceptors (0 - 2) Aliphatic (0, 1) Aromatic (0, 1) Charge (-1, 0, 1) 4. Compute a distance between 2 sites by measuring a normalized distance between 2 fingerprints. 5. Align both targets by exhaustive rotation/translation of one sphere in its site to optimize the similarity score of their fingerprints.

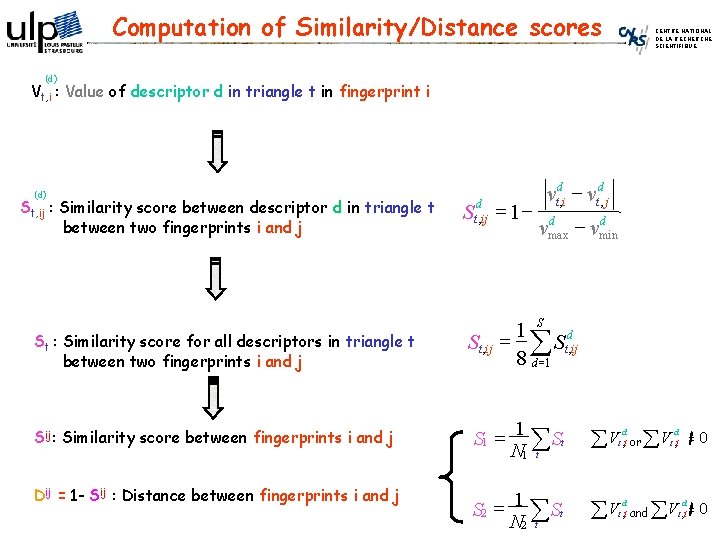
Computation of Similarity/Distance scores CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE (d) Vt, i : Value of descriptor d in triangle t in fingerprint i (d) St, ij : Similarity score between descriptor d in triangle t between two fingerprints i and j St : Similarity score for all descriptors in triangle t between two fingerprints i and j Sij: Similarity score between fingerprints i and j Dij = 1 - Sij : Distance between fingerprints i and j Std, ij = 1 - vtd, i - vtd, j d d - vmin vmax 1 S d St , ij = å St , ij 8 d =1 S 1 = 1 åSt N 1 t åV or S 2 = 1 åSt N 2 t åV and d t , i åV d t , i =0 åV 0 d t , i =

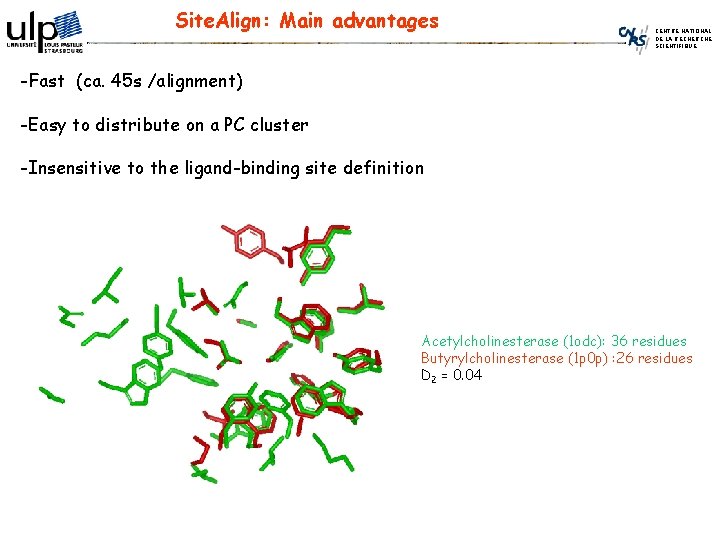
Site. Align: Main advantages CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE -Fast (ca. 45 s /alignment) -Easy to distribute on a PC cluster -Insensitive to the ligand-binding site definition Acetylcholinesterase (1 odc): 36 residues Butyrylcholinesterase (1 p 0 p) : 26 residues D 2 = 0. 04

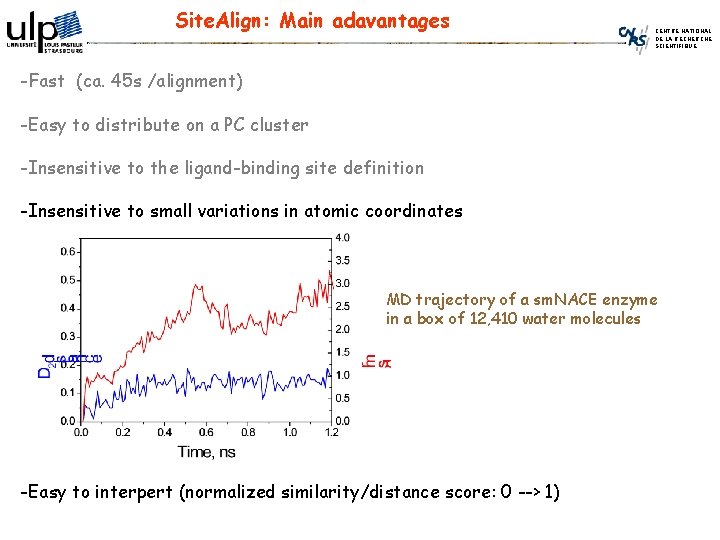
Site. Align: Main adavantages CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE -Fast (ca. 45 s /alignment) -Easy to distribute on a PC cluster -Insensitive to the ligand-binding site definition -Insensitive to small variations in atomic coordinates MD trajectory of a sm. NACE enzyme in a box of 12, 410 water molecules -Easy to interpert (normalized similarity/distance score: 0 --> 1)

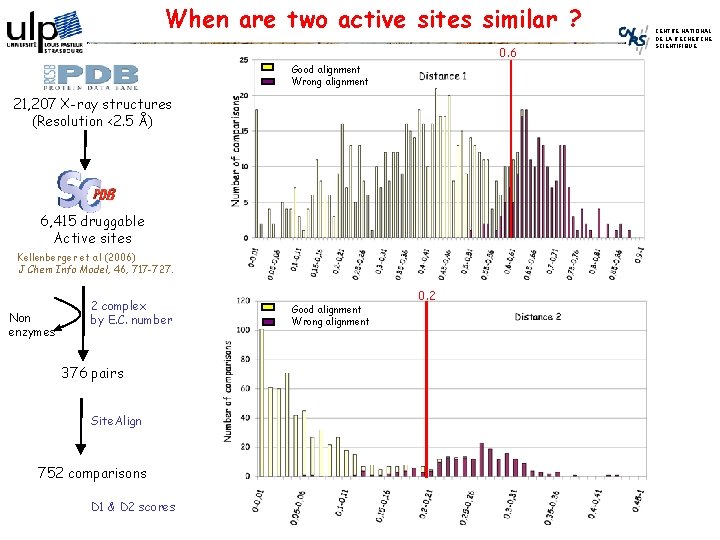
When are two active sites similar ? 0. 6 Good alignment Wrong alignment 21, 207 X-ray structures (Resolution <2. 5 Å) 6, 415 druggable Active sites Kellenberger et al (2006) J Chem Info Model, 46, 717 -727. Non enzymes 2 complex by E. C. number 376 pairs Site. Align 752 comparisons D 1 & D 2 scores Good alignment Wrong alignment 0. 2 CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE

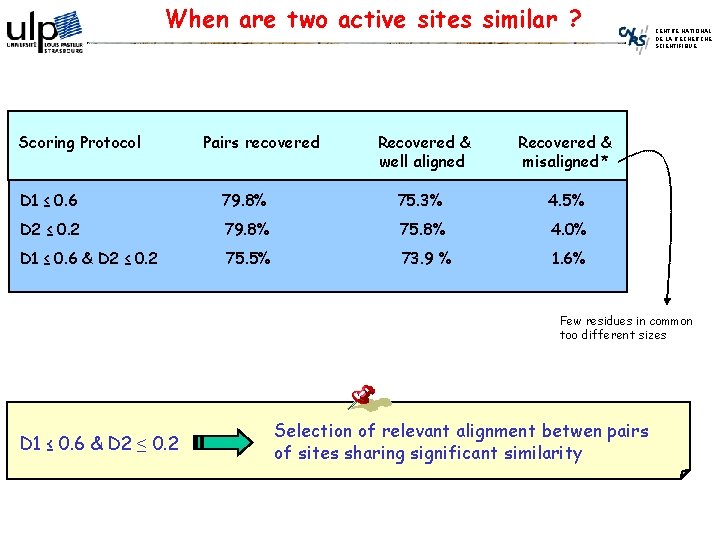
When are two active sites similar ? Scoring Protocol Pairs recovered Recovered & well aligned Recovered & misaligned* D 1 ≤ 0. 6 79. 8% 75. 3% 4. 5% D 2 ≤ 0. 2 79. 8% 75. 8% 4. 0% D 1 ≤ 0. 6 & D 2 ≤ 0. 2 75. 5% 73. 9 % 1. 6% CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE Few residues in common too different sizes D 1 ≤ 0. 6 & D 2 ≤ 0. 2 Selection of relevant alignment betwen pairs of sites sharing significant similarity

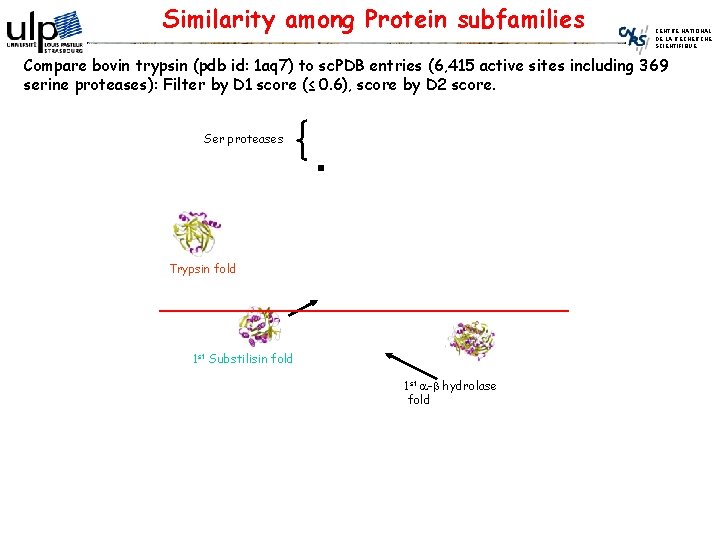
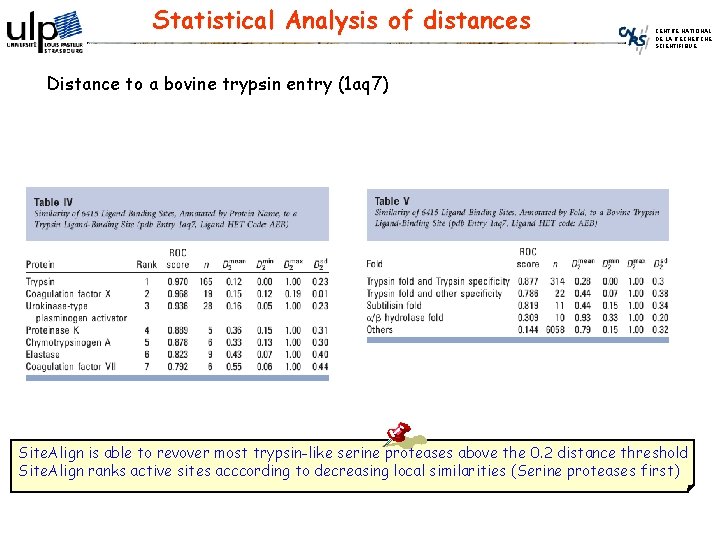
Similarity among Protein subfamilies CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE Compare bovin trypsin (pdb id: 1 aq 7) to sc. PDB entries (6, 415 active sites including 369 serine proteases): Filter by D 1 score (≤ 0. 6), score by D 2 score. Ser proteases Trypsin fold 1 st Substilisin fold 1 st a-b hydrolase fold

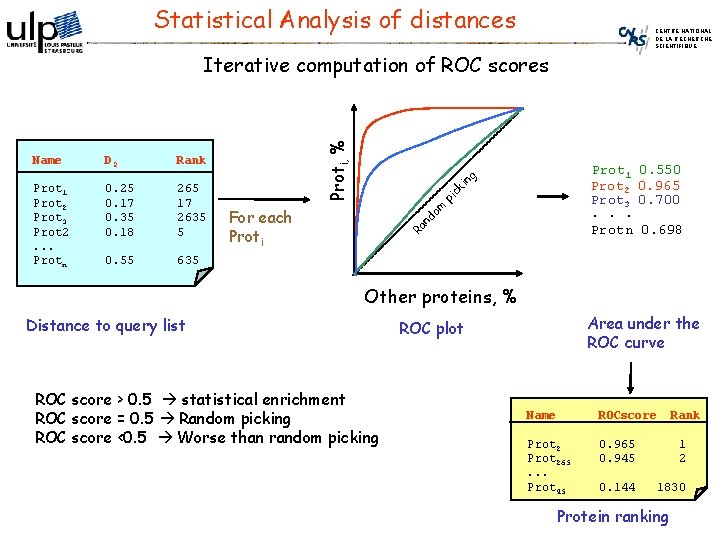
Statistical Analysis of distances CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE 265 17 2635 5 0. 55 635 ck in g 0. 25 0. 17 0. 35 0. 18 pi Prot 1 Prot 2 Prot 3 Prot 2. . . Protn Prot 1 Prot 2 Prot 3. . . Protn om Rank For each Proti nd D 2 Ra Name Proti, % Iterative computation of ROC scores 0. 550 0. 965 0. 700 0. 698 Other proteins, % Distance to query list ROC score > 0. 5 statistical enrichment ROC score = 0. 5 Random picking ROC score <0. 5 Worse than random picking Area under the ROC curve ROC plot Name ROCscore Rank Prot 265. . . Prot 45 0. 965 0. 945 1 2 0. 144 1830 Protein ranking

Statistical Analysis of distances CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE Distance to a bovine trypsin entry (1 aq 7) Site. Align is able to revover most trypsin-like serine proteases above the 0. 2 distance threshold Site. Align ranks active sites acccording to decreasing local similarities (Serine proteases first)

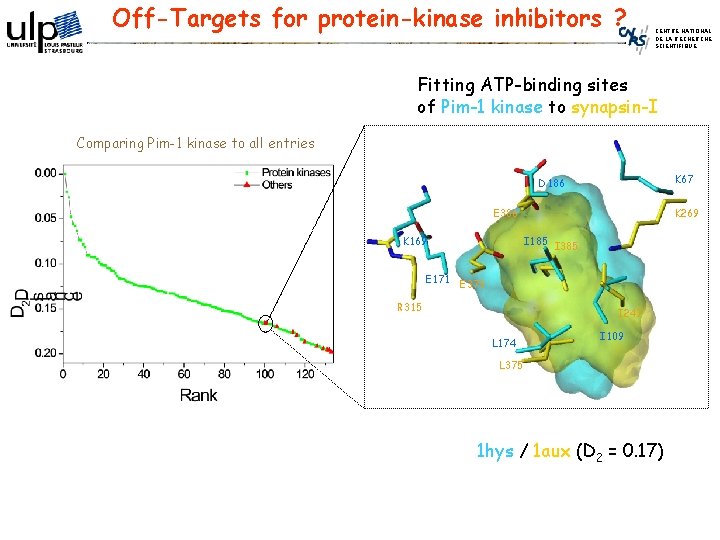
Off-Targets for protein-kinase inhibitors ? CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE Fitting ATP-binding sites of Pim-1 kinase to synapsin-I Comparing Pim-1 kinase to all entries K 67 D 186 E 386 K 169 K 269 I 185 I 385 E 171 E 373 R 315 I 247 L 174 I 109 L 375 1 hys / 1 aux (D 2 = 0. 17)

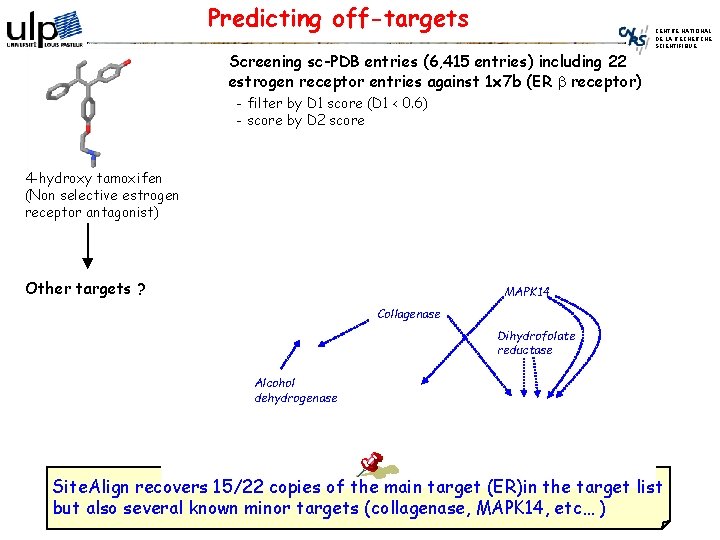
Predicting off-targets CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE Screening sc-PDB entries (6, 415 entries) including 22 estrogen receptor entries against 1 x 7 b (ER b receptor) - filter by D 1 score (D 1 < 0. 6) - score by D 2 score 4 -hydroxy tamoxifen (Non selective estrogen receptor antagonist) Other targets ? MAPK 14 Collagenase Dihydrofolate reductase Alcohol dehydrogenase Site. Align recovers 15/22 copies of the main target (ER)in the target list but also several known minor targets (collagenase, MAPK 14, etc… )

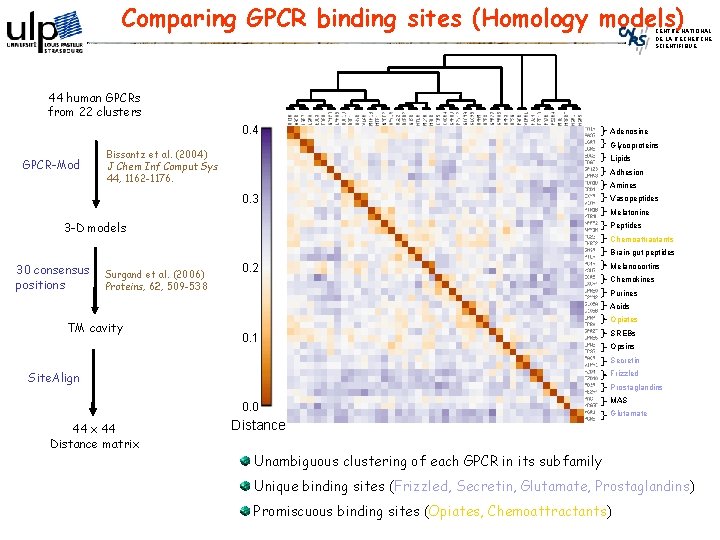
Comparing GPCR binding sites (Homology models) CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE 44 human GPCRs from 22 clusters 0. 4 GPCR-Mod Adenosine Glycoproteins Bissantz et al. (2004) J Chem Inf Comput Sys 44, 1162 -1176. Lipids Adhesion Amines 0. 3 Vasopeptides Melatonine 3 -D models Peptides Chemoattractants Brain-gut peptides 30 consensus positions Surgand et al. (2006) Proteins, 62, 509 -538 0. 2 Melanocortins Chemokines Purines Acids TM cavity Opiates 0. 1 SREBs Opsins Secretin Frizzled Site. Align Prostaglandins 0. 0 44 x 44 Distance matrix Distance MAS Glutamate Unambiguous clustering of each GPCR in its subfamily Unique binding sites (Frizzled, Secretin, Glutamate, Prostaglandins) Promiscuous binding sites (Opiates, Chemoattractants)

Conclusions CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE Binding site comparisons is an emerging tool for : - the annotation of genomic protein structures - the biological profiling of bioactive compounds Chemogenomic paradigm Academia has the time to develop new mining tools but lack high-quality and homogeneous data (binding matrices) Industry has plenty of these data, fewer time to mine it, but does not share it with the academia

Why computing @ IN 2 P 3 ? CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE National Computing centers (CINES, IDRIS) unsuited to our needs in terms of: - Architecture and OS - Computing power A full matrix (1500 x 1500 proteins) requires ca. 40 K cpu hours - Constrained ressource allocation procedure IN 2 P 3 facilities are much more flexible: - Architecture and OS similar to in-house (Intel PC/Linux) - No need to use PVM/MPI/Open. MP libraries for parallel computing - Simple parallelisation of input data (one job – one proc) - No formal ressources allocation request - Enough computing power to compute full similarity matrices

Acknowledgments CENTRE NATIONAL DE LA RECHERCHE SCIENTIFIQUE sc-PDB development Dr. Esther Kellenberger Nicodème Paul Pascal Müller Site. Align: Development and Validation Jean-Sébastien Surgand Claire Schalon Guillaume Bret Nicolas Foata