2 Ponds which were previously positive were sampled











- Slides: 25

• 2 Ponds which were previously positive were sampled in the North Qld farm . (pos on 31/10/15) • 980 prawns selected from 98 ponds on 13 Farms. • There were no detections of the pir. A toxin gene by q. PCR. Indicates positive locations from OIE notification 196695


The farmers guide to accepting real time polyermase chain Reaction (q. PCR) results : is that Cost? What “If you think the cost of Direct $ to fix an incorrect result understanding is or false positive) (falseq. PCR negative excessive you may pay the price of its absence” Or even worse you may make long term investment decisions based on incorrect results and then additionally need to direct $ on what should have been done in the first place (over or underestimating the importance of an agent in genetic selection programs by using the wrong assays)

Results Target What did you look for ? Assay How well does that test work ? What is in place to monitor the test is working? Sample What samples were used? How well did that work ? Testing process

Results Target What was not detected?

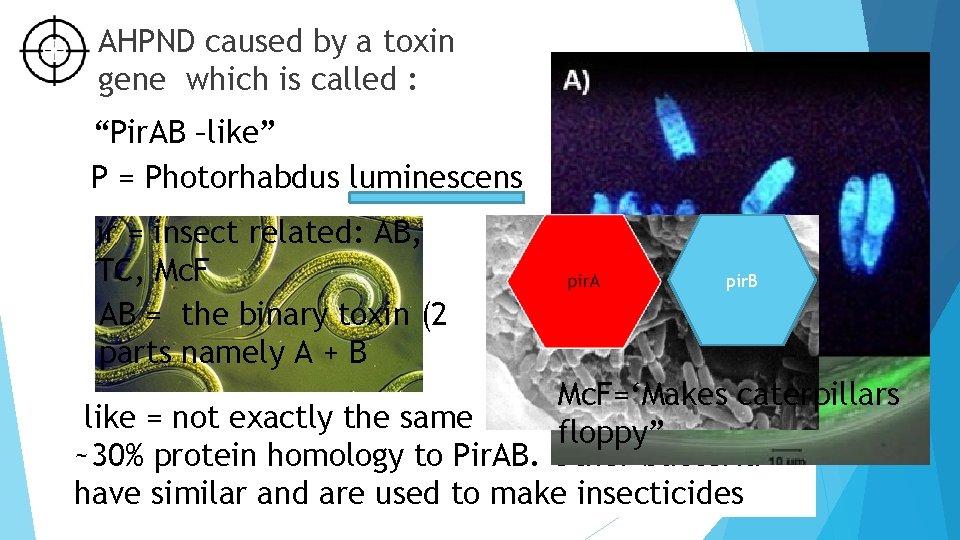
AHPND caused by a toxin gene which is called : “Pir. AB –like” P = Photorhabdus luminescens ir = insect related: AB, TC, Mc. F AB = the binary toxin (2 parts namely A + B pir. B Mc. F=‘Makes caterpillars like = not exactly the same floppy” ~30% protein homology to Pir. AB. Other bacteria have similar and are used to make insecticides

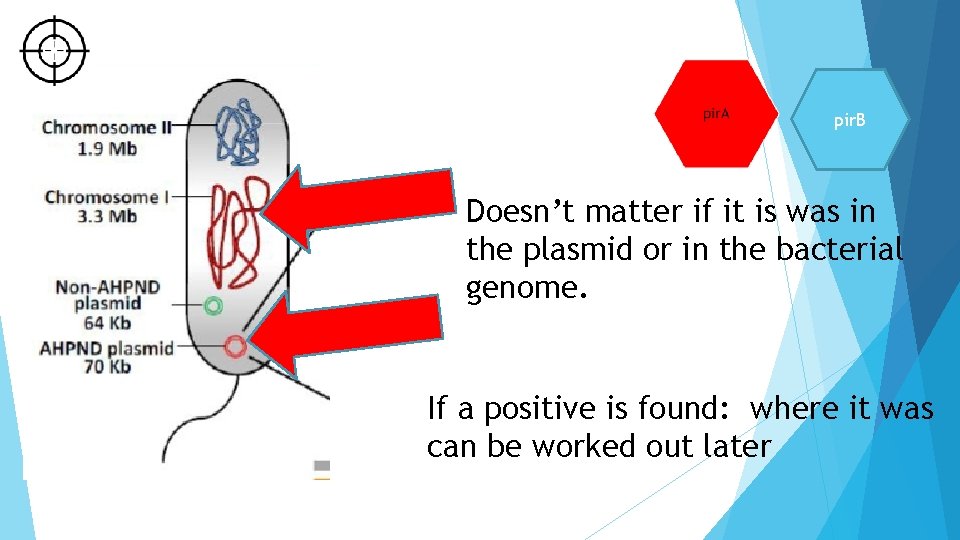
pir. B Doesn’t matter if it is was in the plasmid or in the bacterial genome. If a positive is found: where it was can be worked out later

Assay

Pir copies/u. L 10 1000000 Neg 100 10 1000000 10 Neg 10 1000000 Ct AP 34. 26 16. 83 Undetected 30. 37 33. 86 16. 81 Undetected 30. 42 33. 81 16. 94 33. 51 Undetected 33. 61 16. 8 15 samples

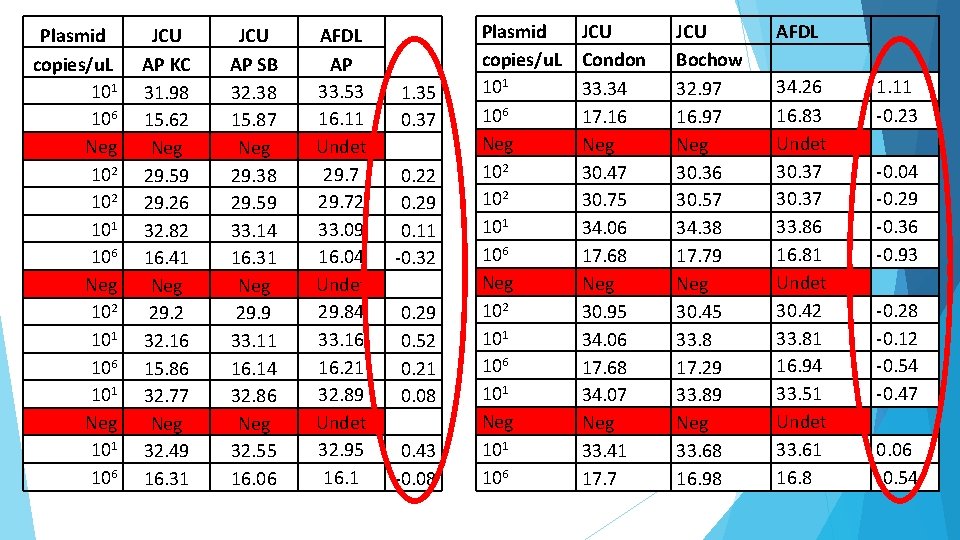
Plasmid copies/u. L 101 106 Neg 102 101 106 101 Neg 101 106 JCU AP KC 31. 98 15. 62 Neg 29. 59 29. 26 32. 82 16. 41 Neg 29. 2 32. 16 15. 86 32. 77 Neg 32. 49 16. 31 JCU AP SB 32. 38 15. 87 Neg 29. 38 29. 59 33. 14 16. 31 Neg 29. 9 33. 11 16. 14 32. 86 Neg 32. 55 16. 06 AFDL AP 33. 53 16. 11 Undet 29. 72 33. 09 16. 04 Undet 29. 84 33. 16 16. 21 32. 89 Undet 32. 95 16. 1 1. 35 0. 37 0. 22 0. 29 0. 11 -0. 32 0. 29 0. 52 0. 21 0. 08 0. 43 -0. 08 Plasmid copies/u. L 101 106 Neg 102 101 106 101 Neg 101 106 JCU Condon 33. 34 17. 16 Neg 30. 47 30. 75 34. 06 17. 68 Neg 30. 95 34. 06 17. 68 34. 07 Neg 33. 41 17. 7 JCU Bochow 32. 97 16. 97 Neg 30. 36 30. 57 34. 38 17. 79 Neg 30. 45 33. 8 17. 29 33. 89 Neg 33. 68 16. 98 AFDL 34. 26 16. 83 Undet 30. 37 33. 86 16. 81 Undet 30. 42 33. 81 16. 94 33. 51 Undet 33. 61 16. 8 1. 11 -0. 23 -0. 04 -0. 29 -0. 36 -0. 93 -0. 28 -0. 12 -0. 54 -0. 47 0. 06 -0. 54

Sample • Needed to increase the bacterial numbers present in samples • Confirm the presence of bacteria in the samples • Have samples in formats that would allow bacterial cultures to be grown if there was a positive detection. • Have samples in formats that could be sent to BQ and AFDL for repeat testing

Sample

Results Target What did you look for ? Assay How well does that test work ? What is in place to monitor the test is working? Sample What samples were used? How well did that work ? Testing process

Results Does the assay to detect the pir. A toxin gene perform consistently, accurately and specifically to detect the pir. A gene ? Test performance

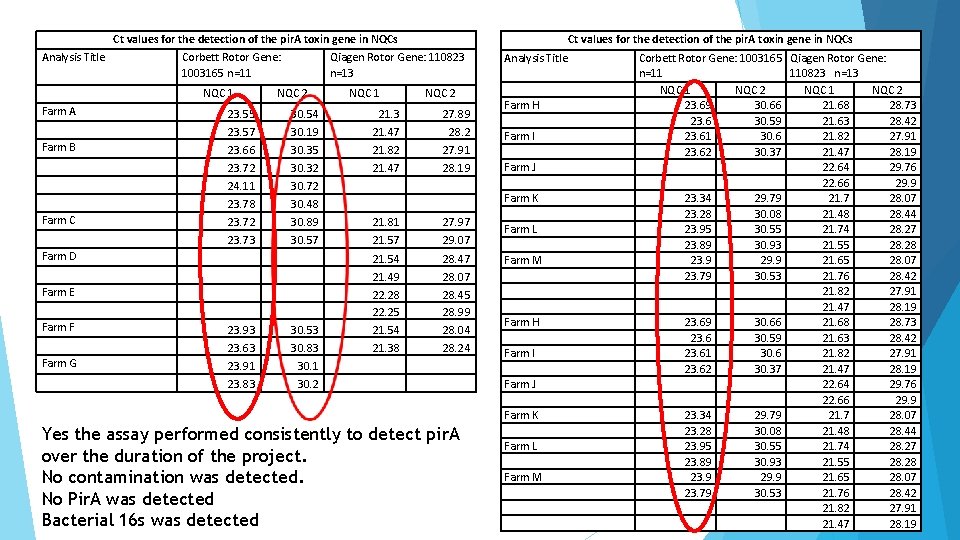
Ct values for the detection of the pir. A toxin gene in NQCs Analysis Title Corbett Rotor Gene: 1003165 n=11 NQC 1 Qiagen Rotor Gene: 110823 n=13 NQC 2 NQC 1 NQC 2 Farm A 23. 55 30. 54 21. 3 27. 89 23. 57 30. 19 21. 47 28. 2 Farm B 23. 66 30. 35 21. 82 27. 91 23. 72 30. 32 24. 11 Farm C 21. 47 28. 19 30. 72 23. 78 30. 48 23. 72 30. 89 21. 81 27. 97 23. 73 29. 07 30. 57 21. 57 Farm D 21. 54 28. 47 21. 49 28. 07 Farm E Farm F 22. 28 22. 25 28. 45 28. 99 23. 93 30. 53 21. 54 28. 04 23. 63 30. 83 21. 38 28. 24 Farm G 23. 91 30. 1 23. 83 30. 2 Yes the assay performed consistently to detect pir. A over the duration of the project. No contamination was detected. No Pir. A was detected Bacterial 16 s was detected Analysis Title Farm H Farm I Farm J Farm K Farm L Farm M Corbett Rotor Gene: 1003165 Qiagen Rotor Gene: n=11 110823 n=13 NQC 1 NQC 2 23. 69 30. 66 21. 68 28. 73 23. 6 30. 59 21. 63 28. 42 23. 61 30. 6 21. 82 27. 91 23. 62 30. 37 21. 47 28. 19 22. 64 29. 76 22. 66 29. 9 23. 34 29. 79 21. 7 28. 07 23. 28 30. 08 21. 48 28. 44 23. 95 30. 55 21. 74 28. 27 23. 89 30. 93 21. 55 28. 28 23. 9 29. 9 21. 65 28. 07 23. 79 30. 53 21. 76 28. 42 21. 82 27. 91 21. 47 28. 19

Thank you: Biosecurity QLD: without their consent we could not have been involved in this project. FRDC: funding to APFA: funding to JCU staff, travel, sample collection and reagents for testing. Helen Jenkins for logistical arrangements and purchasing. AFDL AAHL (CSIRO): Method instructions, positive controls, equivalence testing panel. (Nick Moody, David Cummins, Lynette Williams, John Hoad). JCU Townsville: In kind access to equipment and laboratories (College of Public Health, Medical and Veterinary Sciences) Sampling Teams: Pro. Aqua: Alistair Dick Ridley Australia: Matt Briggs and Caitlin Riguet BIARC: Brian Paterson JCU: Shaun Bochow, Kelly Condon APFs that hosted our sampling teams: Australian Prawn Farms, Seafarm, Tru Blu, Gold Coast Marine Aquaculture, Pacific Reef, Rocky Point Prawn Farm, Truloff Fresh Prawns, David Lin, Campwin Beach Prawn Farm, GI Rural Pty Ltd. , Paradise Prawn Farm.

Questions ?



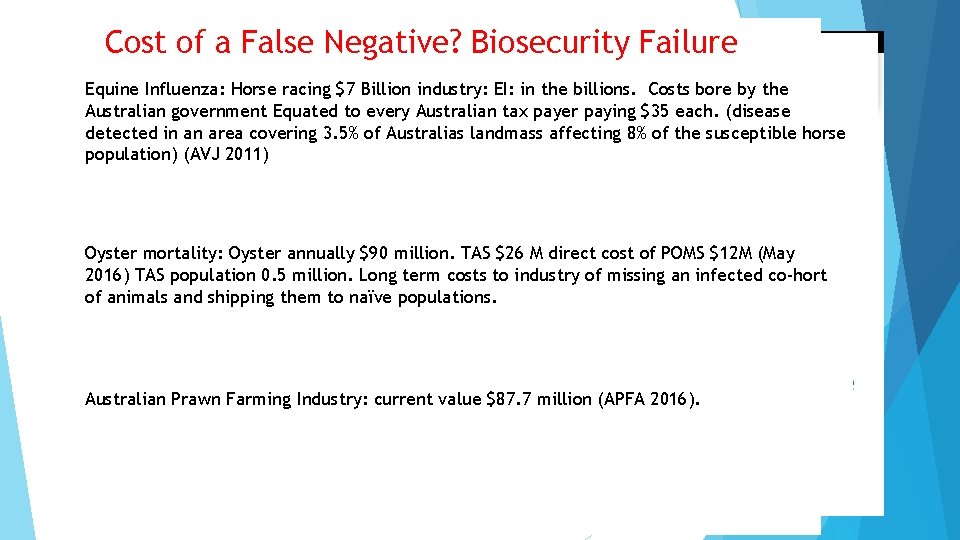
Cost of a False Negative? Biosecurity Failure Equine Influenza: Horse racing $7 Billion industry: EI: in the billions. Costs bore by the Australian government Equated to every Australian tax payer paying $35 each. (disease detected in an area covering 3. 5% of Australias landmass affecting 8% of the susceptible horse population) (AVJ 2011) Cost of a False Oyster mortality: Oyster annually $90 million. TAS $26 Positive: M direct cost of POMS $12 M (May 2016) TAS population 0. 5 million. Long term costs to industry of missing an infected co-hort of animals and shipping them to naïve populations. Australian Prawn Farming Industry: current value $87. 7 million (APFA 2016).


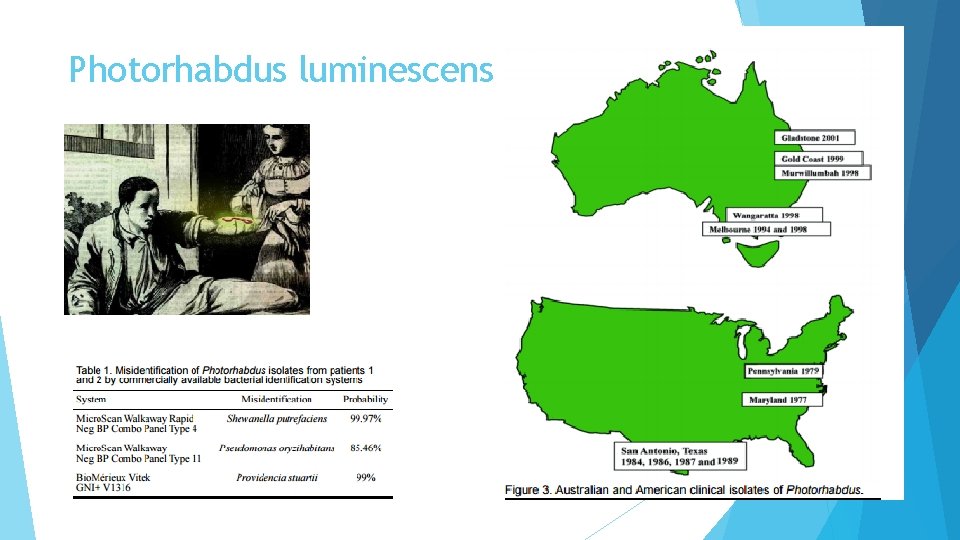
Photorhabdus luminescens


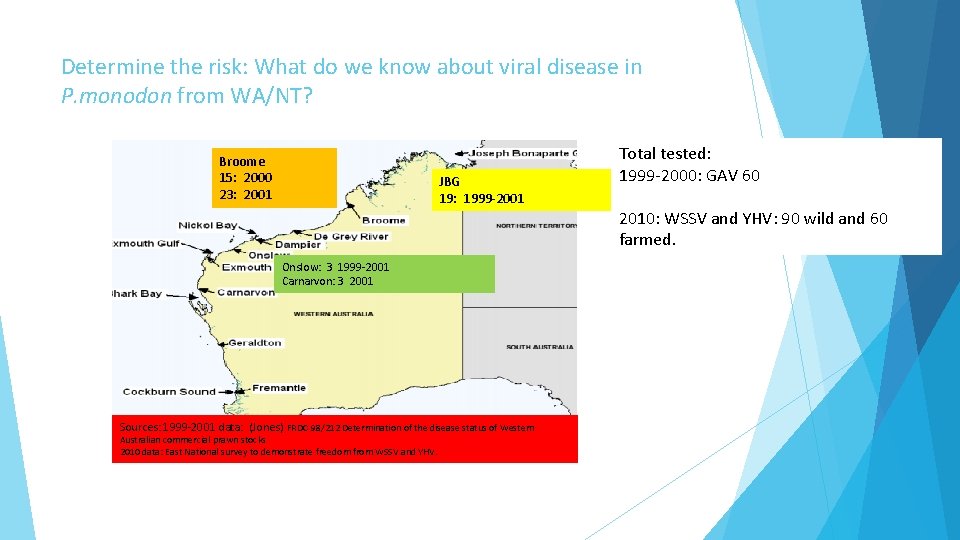
Determine the risk: What do we know about viral disease in P. monodon from WA/NT? Broome 15: 2000 23: 2001 JBG 19: 1999 -2001 Onslow: 3 1999 -2001 Carnarvon: 3 2001 Sources: 1999 -2001 data: (Jones) FRDC 98/212 Determination of the disease status of Western Australian commercial prawn stocks 2010 data: East National survey to demonstrate freedom from WSSV and YHV. Total tested: 1999 -2000: GAV 60 2010: WSSV and YHV: 90 wild and 60 farmed.

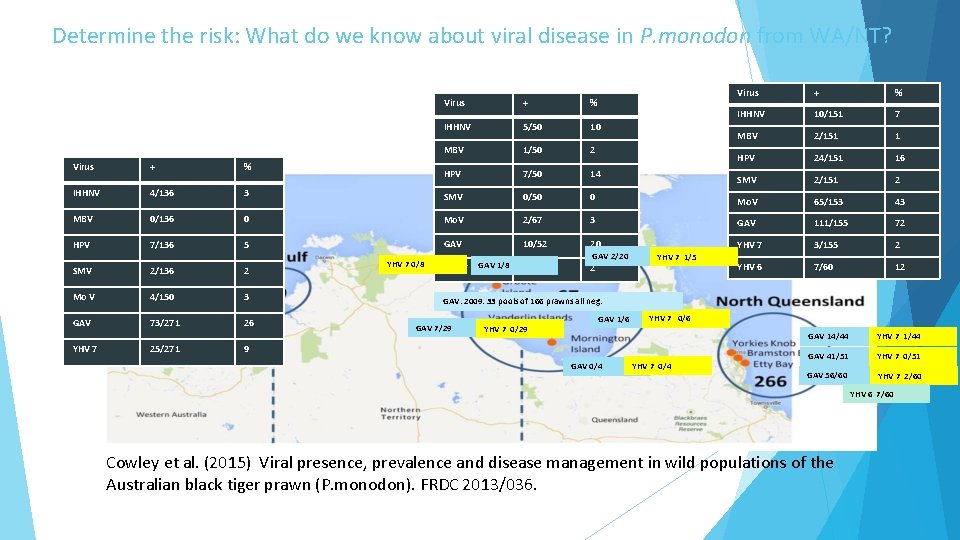
Determine the risk: What do we know about viral disease in P. monodon from WA/NT? Virus + % IHHNV 5/50 10 MBV 1/50 2 HPV 7/50 14 Virus + % IHHNV 10/151 7 MBV 2/151 1 HPV 24/151 16 SMV 2/151 2 IHHNV 4/136 3 SMV 0/50 0 Mo. V 65/153 43 MBV 0/136 0 Mo. V 2/67 3 GAV 111/155 72 HPV 7/136 5 GAV 10/52 20 YHV 7 3/155 2 YHV 6 7/60 12 SMV 2/136 2 Mo V 4/150 3 GAV 73/271 26 YHV 7 25/271 YHV 7 0/8 YHV 7 GAV 1/8 GAV 2/20 1/52 2 YHV 7 1/5 GAV: 2009: 33 pools of 166 prawns all neg. GAV 7/29 YHV 7 0/29 GAV 1/6 YHV 7 0/6 9 GAV 0/4 YHV 7 0/4 GAV 14/44 YHV 7 1/44 GAV 41/51 YHV 7 0/51 GAV 56/60 YHV 7 2/60 YHV 6 7/60 Cowley et al. (2015) Viral presence, prevalence and disease management in wild populations of the Australian black tiger prawn (P. monodon). FRDC 2013/036.