134 Docking placing a ligand into a receptor





- Slides: 34

1/34 Docking: placing a ligand into a receptor cavity Esther Kellenberger Faculté de Pharmacie UMR 7200, Illkirch Tel: 03 90 24 42 21 e-mail: esther. kellenberger@unistra. fr

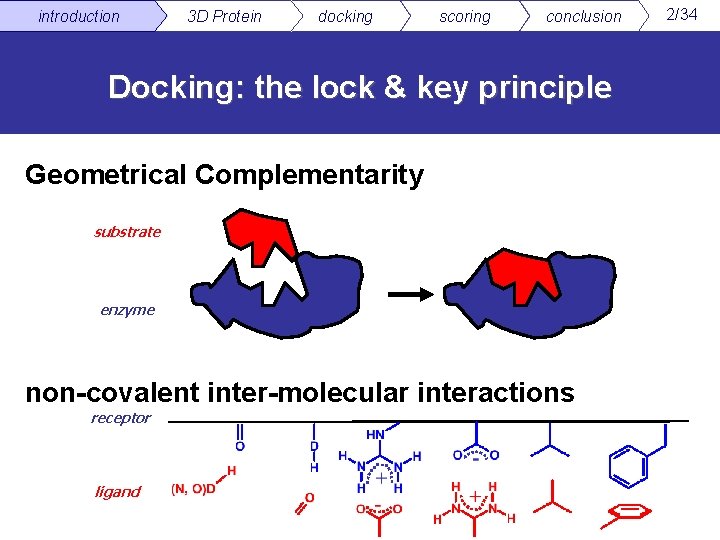
introduction 3 D Protein docking scoring conclusion Docking: the lock & key principle Geometrical Complementarity substrate enzyme non-covalent inter-molecular interactions receptor ligand 2/34

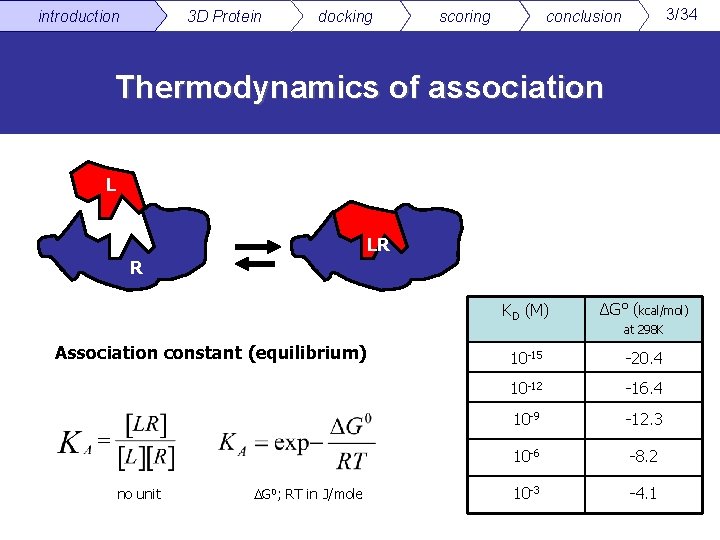
introduction 3 D Protein docking scoring 3/34 conclusion Thermodynamics of association L LR R Association constant (equilibrium) no unit G 0; RT in J/mole KD (M) ΔG° (kcal/mol) 10 -15 -20. 4 10 -12 -16. 4 10 -9 -12. 3 10 -6 -8. 2 10 -3 -4. 1 at 298 K

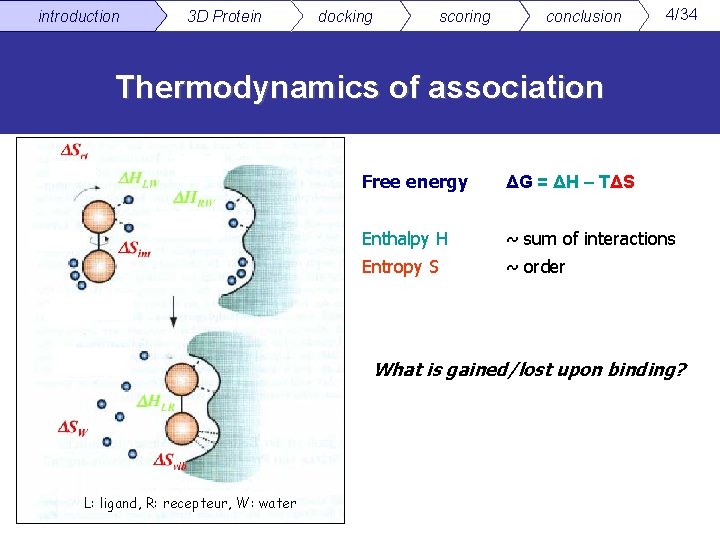
introduction 3 D Protein docking scoring conclusion 4/34 Thermodynamics of association Free energy ΔG = ΔH – TΔS Enthalpy H ~ sum of interactions Entropy S ~ order What is gained/lost upon binding? L: ligand, R: recepteur, W: water

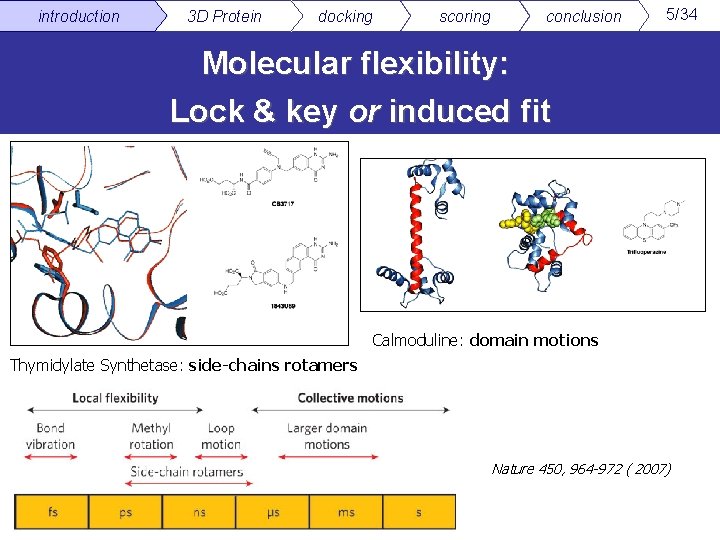
introduction 3 D Protein docking scoring conclusion 5/34 Molecular flexibility: Lock & key or induced fit Calmoduline: domain motions Thymidylate Synthetase: side-chains rotamers Nature 450, 964 -972 ( 2007)

6/34 Chapter 1: Proteins are folded biopolymers

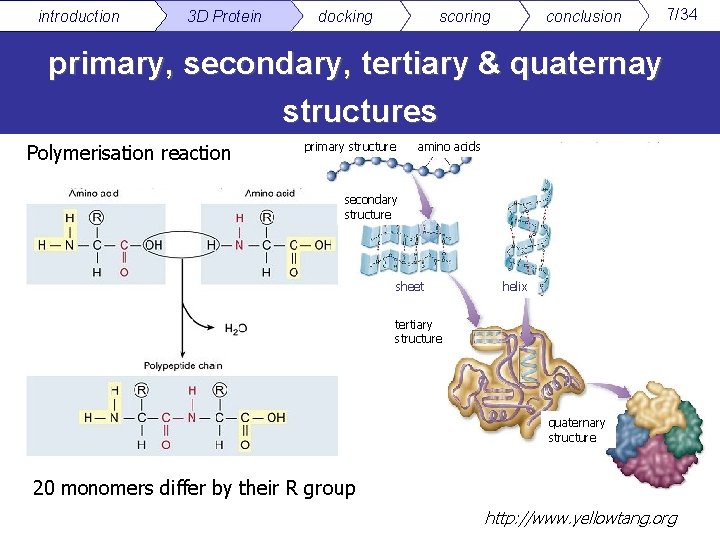
introduction 3 D Protein docking scoring conclusion 7/34 primary, secondary, tertiary & quaternay structures Polymerisation reaction primary structure amino acids secondary structure sheet helix tertiary structure quaternary structure 20 monomers differ by their R group http: //www. yellowtang. org

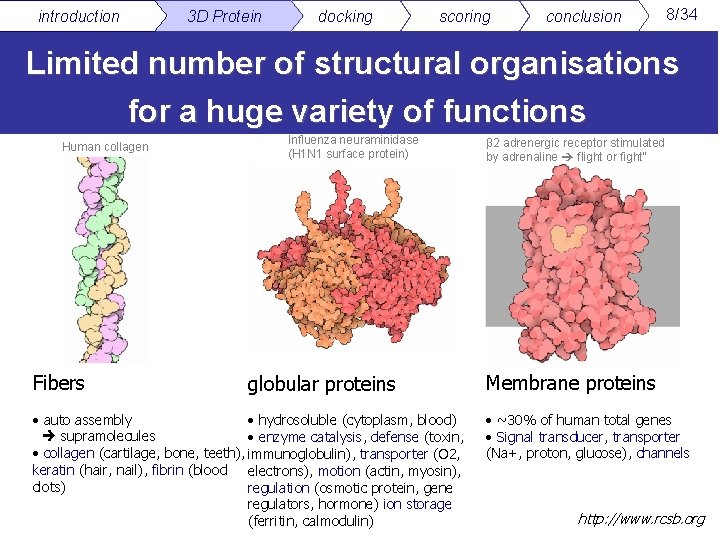
introduction 3 D Protein docking scoring conclusion 8/34 Limited number of structural organisations for a huge variety of functions Human collagen Fibers Influenza neuraminidase (H 1 N 1 surface protein) globular proteins • auto assembly • hydrosoluble (cytoplasm, blood) supramolecules • enzyme catalysis, defense (toxin, • collagen (cartilage, bone, teeth), immunoglobulin), transporter (O 2, keratin (hair, nail), fibrin (blood electrons), motion (actin, myosin), clots) regulation (osmotic protein, gene regulators, hormone) ion storage (ferritin, calmodulin) β 2 adrenergic receptor stimulated by adrenaline flight or fight" Membrane proteins • ~30% of human total genes • Signal transducer, transporter (Na+, proton, glucose), channels http: //www. rcsb. org

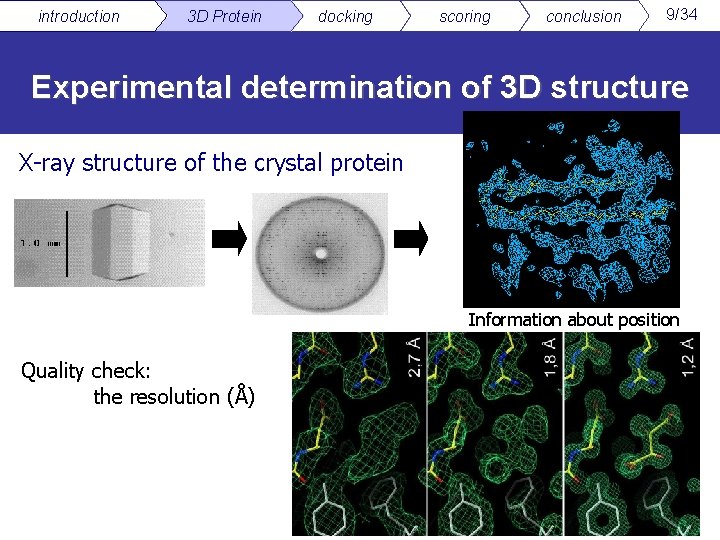
introduction 3 D Protein docking scoring conclusion 9/34 Experimental determination of 3 D structure X-ray structure of the crystal protein Information about position Quality check: the resolution (Å)

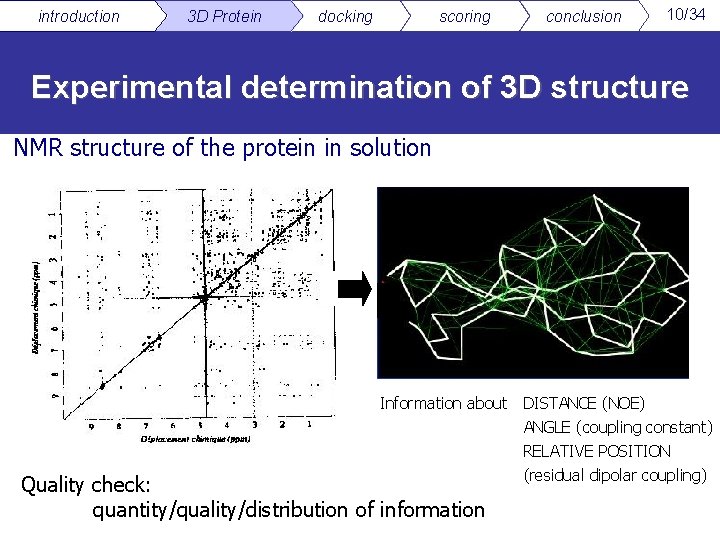
introduction 3 D Protein docking scoring conclusion 10/34 Experimental determination of 3 D structure NMR structure of the protein in solution Information about Quality check: quantity/quality/distribution of information DISTANCE (NOE) ANGLE (coupling constant) RELATIVE POSITION (residual dipolar coupling)

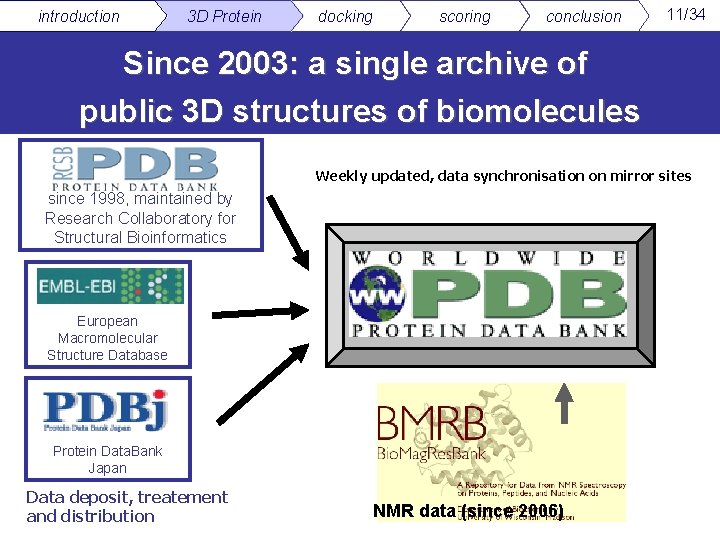
introduction 3 D Protein docking scoring conclusion 11/34 Since 2003: a single archive of public 3 D structures of biomolecules Weekly updated, data synchronisation on mirror sites since 1998, maintained by Research Collaboratory for Structural Bioinformatics European Macromolecular Structure Database Protein Data. Bank Japan Data deposit, treatement and distribution NMR data (since 2006)

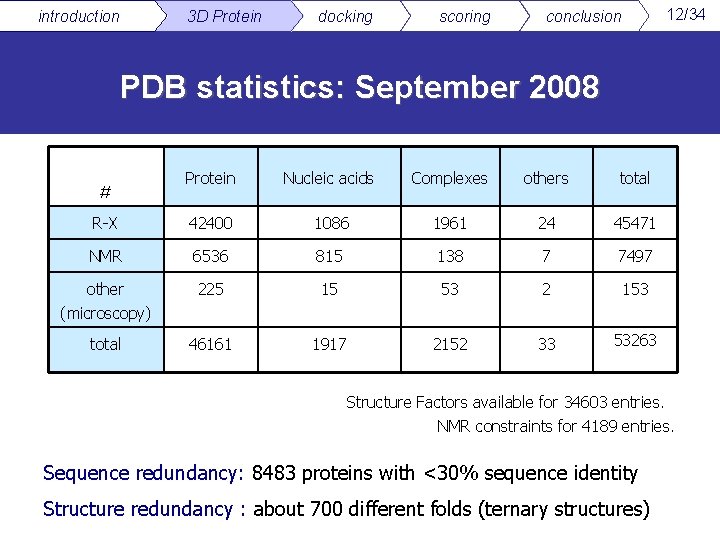
introduction 3 D Protein docking scoring 12/34 conclusion PDB statistics: September 2008 Protein Nucleic acids Complexes others total R-X 42400 1086 1961 24 45471 NMR 6536 815 138 7 7497 other (microscopy) 225 15 53 2 153 total 46161 1917 2152 33 53263 # Structure Factors available for 34603 entries. NMR constraints for 4189 entries. Sequence redundancy: 8483 proteins with <30% sequence identity Structure redundancy : about 700 different folds (ternary structures)

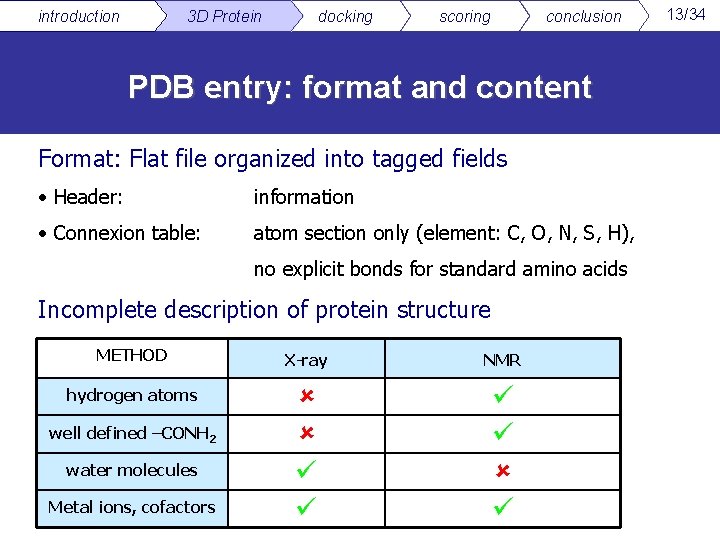
introduction 3 D Protein docking scoring conclusion PDB entry: format and content Format: Flat file organized into tagged fields • Header: information • Connexion table: atom section only (element: C, O, N, S, H), no explicit bonds for standard amino acids Incomplete description of protein structure METHOD X-ray NMR hydrogen atoms well defined –CONH 2 water molecules Metal ions, cofactors 13/34

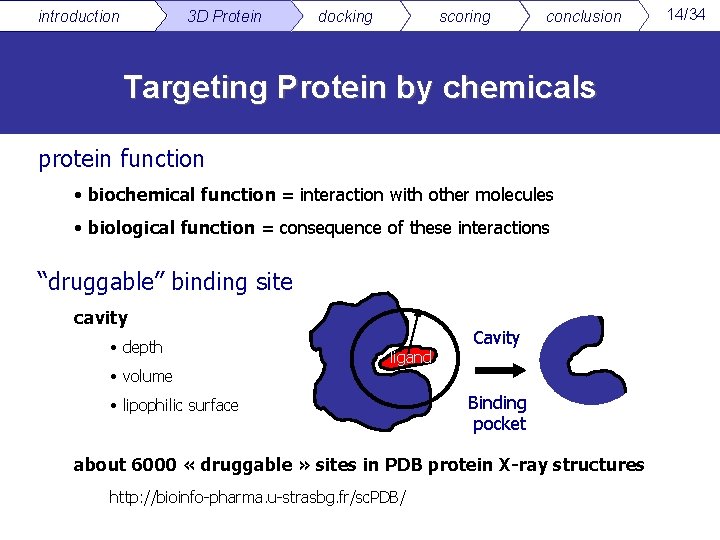
introduction 3 D Protein docking scoring conclusion Targeting Protein by chemicals protein function • biochemical function = interaction with other molecules • biological function = consequence of these interactions “druggable” binding site cavity • depth • volume ligand • lipophilic surface Cavity Binding pocket about 6000 « druggable » sites in PDB protein X-ray structures http: //bioinfo-pharma. u-strasbg. fr/sc. PDB/ 14/34

15/34 Chapter 2: Docking chemicals into protein cavities

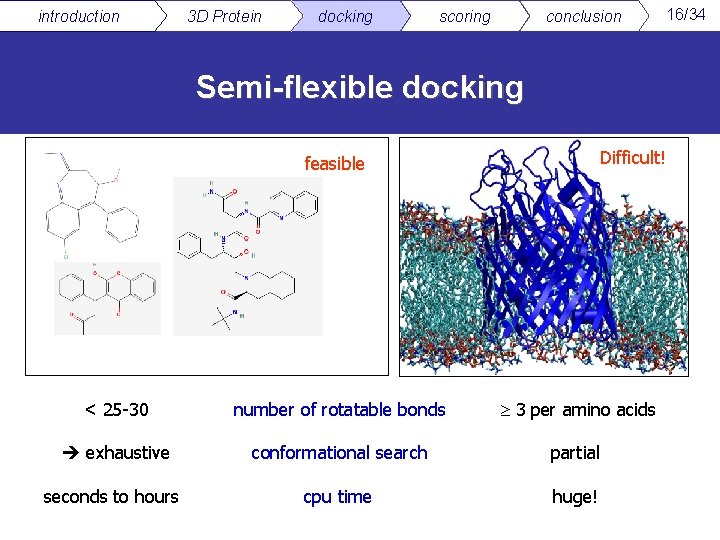
introduction 3 D Protein docking scoring conclusion Semi-flexible docking Difficult! feasible < 25 -30 number of rotatable bonds 3 per amino acids exhaustive conformational search partial seconds to hours cpu time huge! 16/34

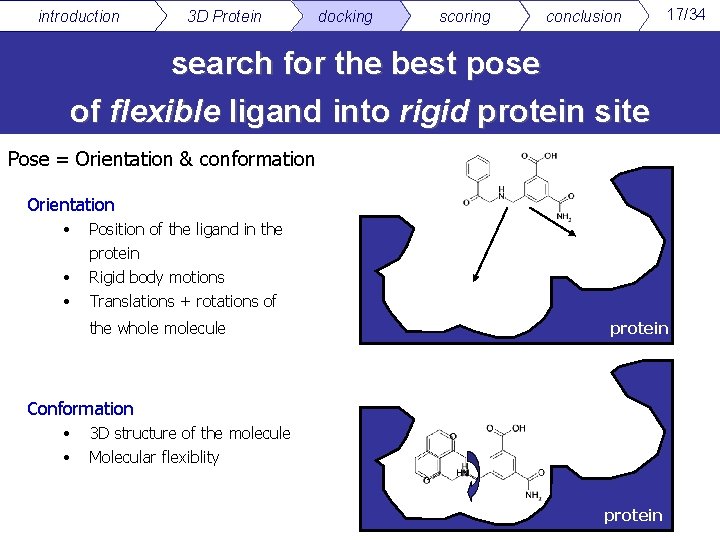
introduction 3 D Protein docking scoring conclusion 17/34 search for the best pose of flexible ligand into rigid protein site Pose = Orientation & conformation Orientation • • • Position of the ligand in the protein Rigid body motions Translations + rotations of the whole molecule protein Conformation • • 3 D structure of the molecule Molecular flexiblity protein

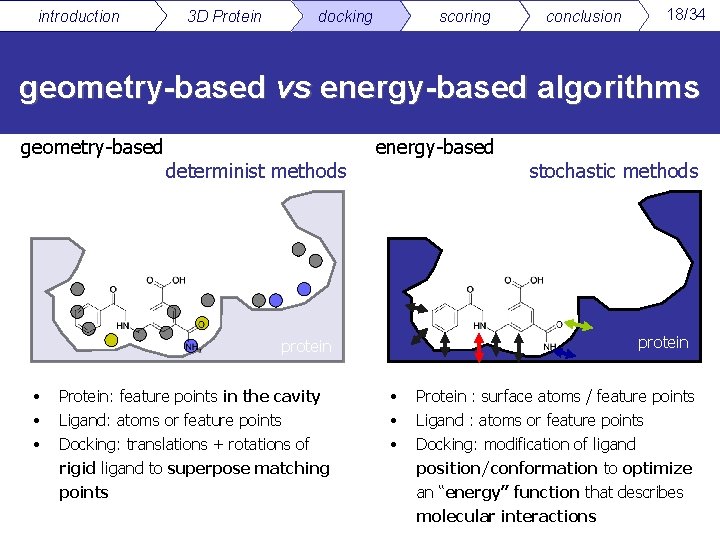
introduction 3 D Protein docking scoring conclusion 18/34 geometry-based vs energy-based algorithms geometry-based determinist methods energy-based protein • • • Protein: feature points in the cavity Ligand: atoms or feature points Docking: translations + rotations of rigid ligand to superpose matching points stochastic methods • • • Protein : surface atoms / feature points Ligand : atoms or feature points Docking: modification of ligand position/conformation to optimize an “energy” function that describes molecular interactions

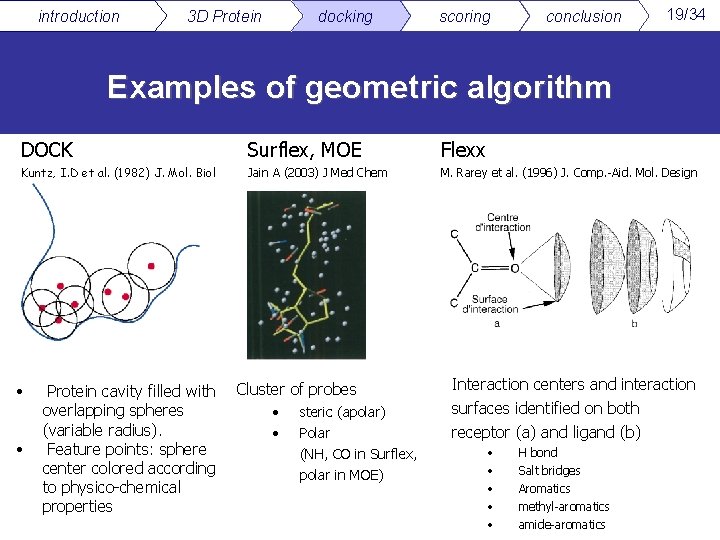
introduction 3 D Protein docking scoring conclusion 19/34 Examples of geometric algorithm DOCK Surflex, MOE Flexx Kuntz, I. D et al. (1982) J. Mol. Biol Jain A (2003) J Med Chem M. Rarey et al. (1996) J. Comp. -Aid. Mol. Design • • Protein cavity filled with overlapping spheres (variable radius). Feature points: sphere center colored according to physico-chemical properties Cluster of probes • • steric (apolar) Polar (NH, CO in Surflex, polar in MOE) Interaction centers and interaction surfaces identified on both receptor (a) and ligand (b) • • • H bond Salt bridges Aromatics methyl-aromatics amide-aromatics

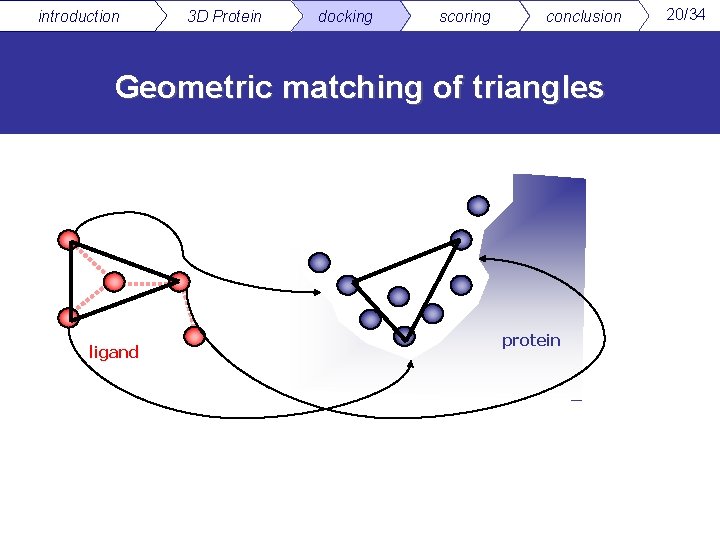
introduction 3 D Protein docking scoring conclusion Geometric matching of triangles ligand protein 20/34

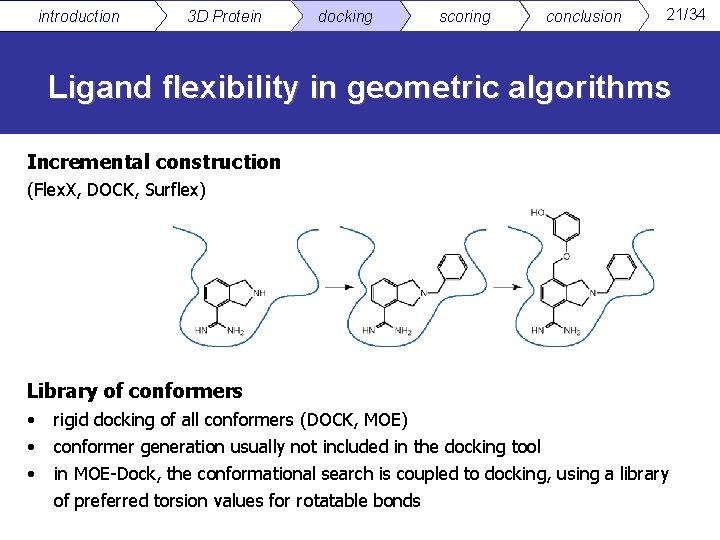
introduction 3 D Protein docking scoring conclusion 21/34 Ligand flexibility in geometric algorithms Incremental construction (Flex. X, DOCK, Surflex) Library of conformers • • • rigid docking of all conformers (DOCK, MOE) conformer generation usually not included in the docking tool in MOE-Dock, the conformational search is coupled to docking, using a library of preferred torsion values for rotatable bonds

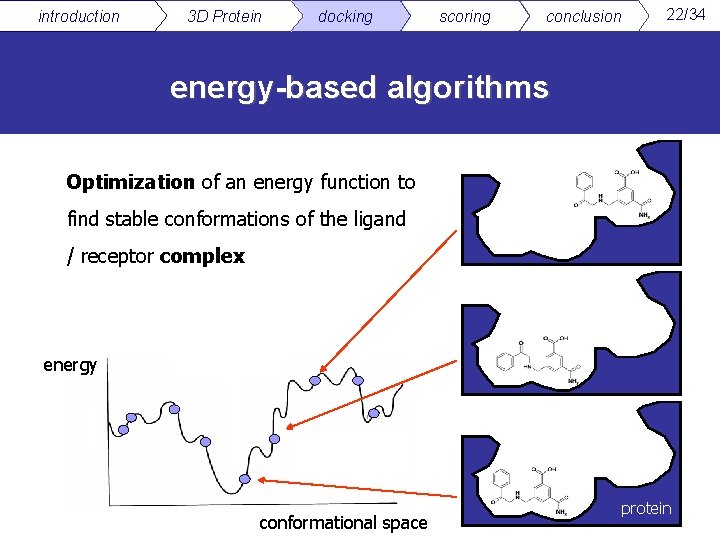
introduction 3 D Protein docking scoring conclusion 22/34 energy-based algorithms Optimization of an energy function to find stable conformations of the ligand / receptor complex energy conformational space protein

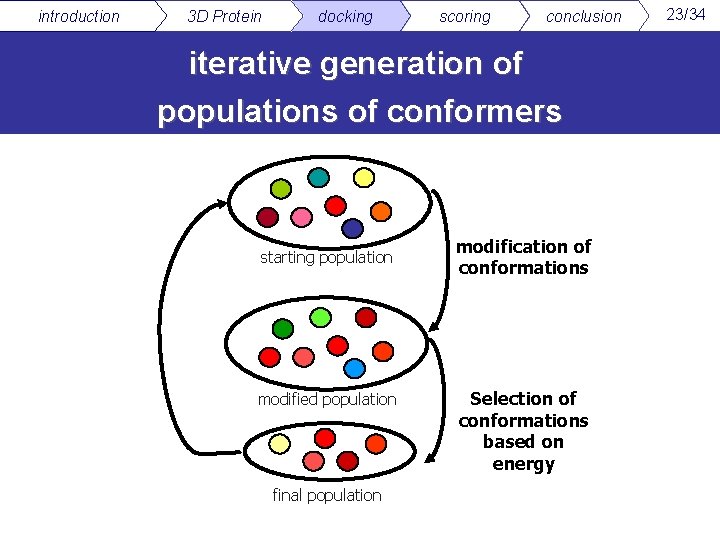
introduction 3 D Protein docking scoring conclusion iterative generation of populations of conformers starting population modified population final population modification of conformations Selection of conformations based on energy 23/34

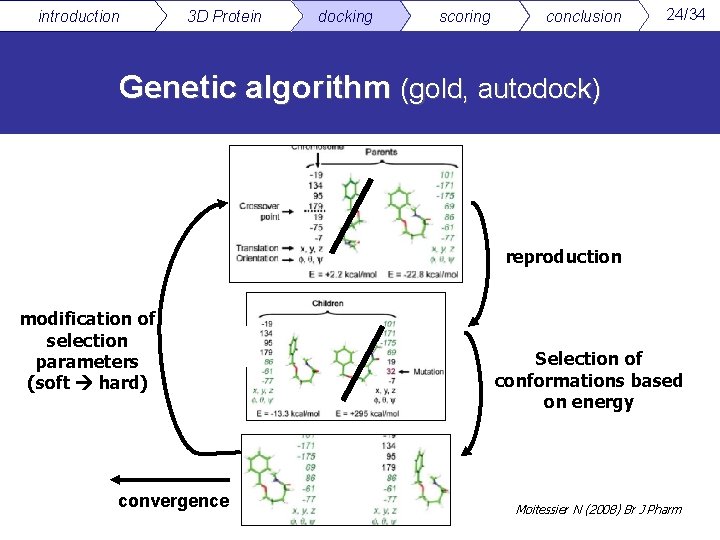
introduction 3 D Protein docking scoring conclusion 24/34 Genetic algorithm (gold, autodock) reproduction modification of selection parameters (soft hard) convergence Selection of conformations based on energy Moitessier N (2008) Br J Pharm

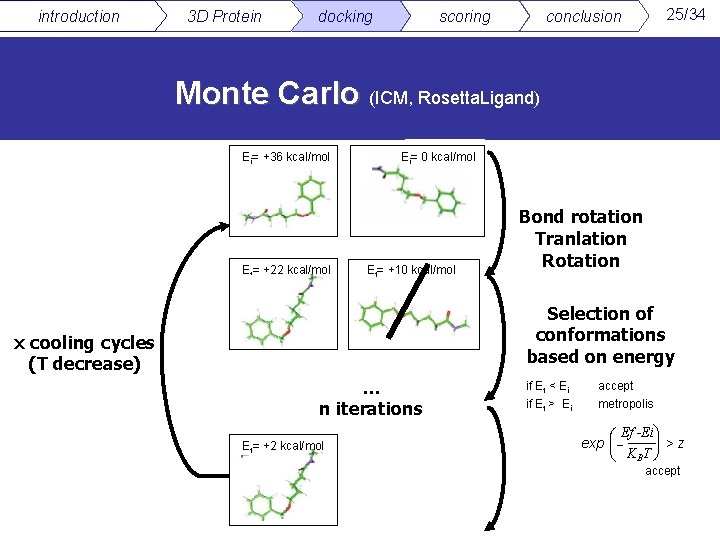
introduction 3 D Protein docking scoring 25/34 conclusion Monte Carlo (ICM, Rosetta. Ligand) Ei= +36 kcal/mol Ef= +22 kcal/mol Ei= 0 kcal/mol Ef= +10 kcal/mol Bond rotation Tranlation Rotation Selection of conformations based on energy x cooling cycles (T decrease) … n iterations Ef= +2 kcal/mol if Ef < Ei if Ef > Ei accept metropolis æ Ef -Ei ö÷ exp çç ÷ >z è K BT ø accept

26/34 Chapter 3: Scoring ligand/receptor interaction

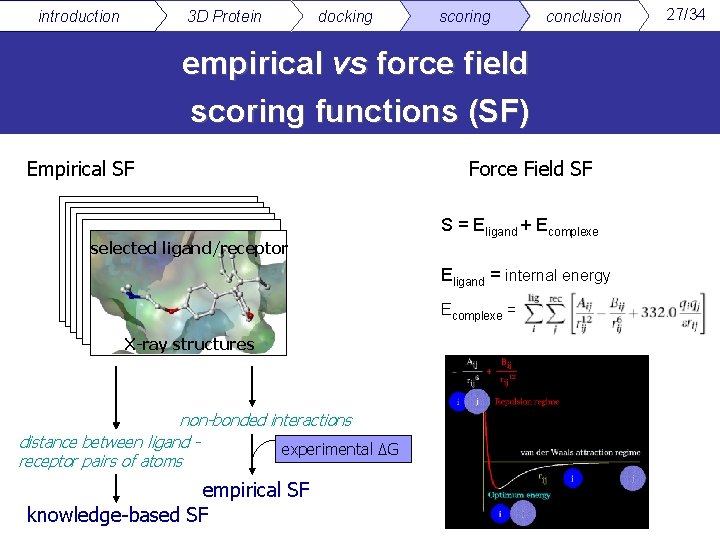
introduction 3 D Protein docking scoring conclusion empirical vs force field scoring functions (SF) Empirical SF selected ligand/receptor Force Field SF S = Eligand + Ecomplexe Eligand = internal energy Ecomplexe = X-ray structures non-bonded interactions distance between ligand experimental ΔG receptor pairs of atoms empirical SF knowledge-based SF 27/34

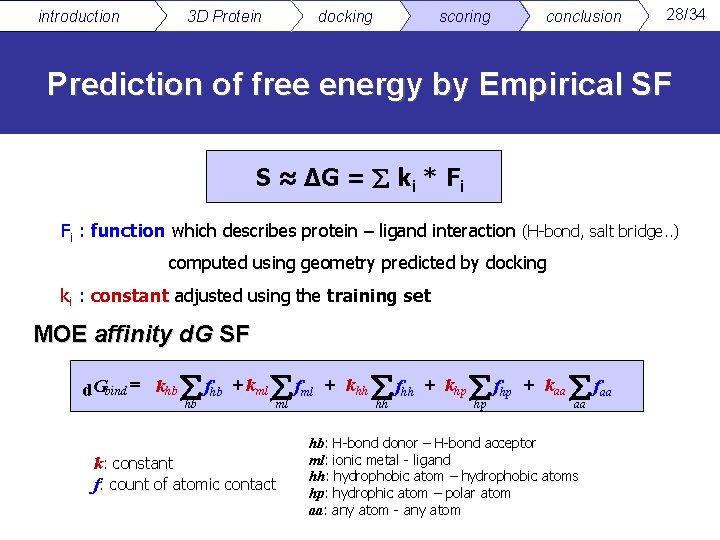
introduction 3 D Protein docking scoring conclusion 28/34 Prediction of free energy by Empirical SF S ≈ ∆G = ki * Fi Fi : function which describes protein – ligand interaction (H-bond, salt bridge. . ) computed using geometry predicted by docking ki : constant adjusted using the training set MOE affinity d. G SF d. Gbind = khb fhb + kml fml + khh fhh + khp fhp + kaa faa hb ml k: constant f: count of atomic contact hh hp aa hb: H-bond donor – H-bond acceptor ml: ionic metal - ligand hh: hydrophobic atom – hydrophobic atoms hp: hydrophic atom – polar atom aa: any atom - any atom

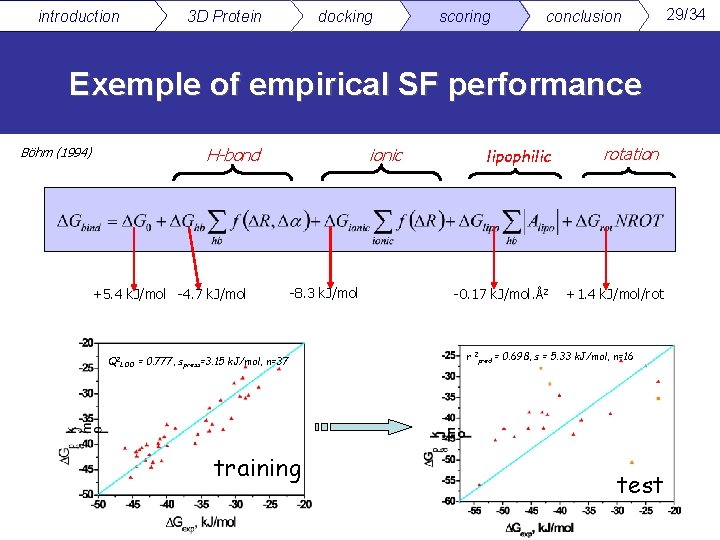
introduction 3 D Protein docking scoring conclusion Exemple of empirical SF performance Böhm (1994) H-bond +5. 4 k. J/mol -4. 7 k. J/mol ionic -8. 3 k. J/mol Q 2 LOO = 0. 777, spress=3. 15 k. J/mol, n=37 training lipophilic -0. 17 k. J/mol. Å2 rotation +1. 4 k. J/mol/rot r 2 pred = 0. 698, s = 5. 33 k. J/mol, n=16 test 29/34

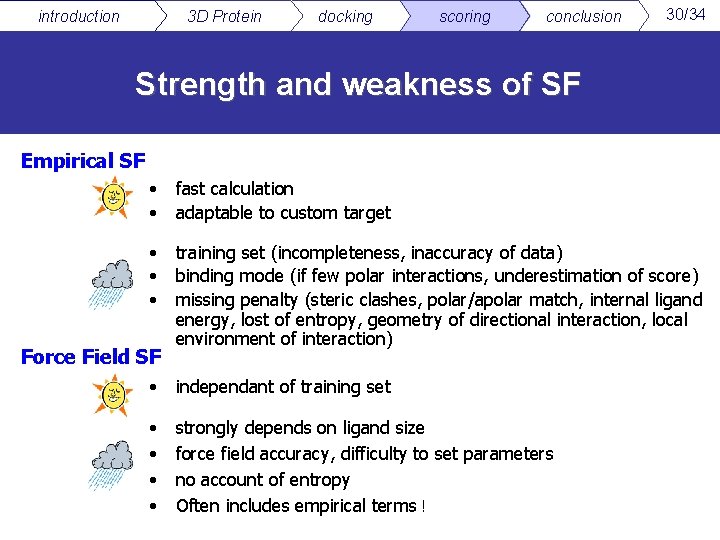
introduction 3 D Protein docking scoring conclusion 30/34 Strength and weakness of SF Empirical SF • • fast calculation adaptable to custom target • • • training set (incompleteness, inaccuracy of data) binding mode (if few polar interactions, underestimation of score) missing penalty (steric clashes, polar/apolar match, internal ligand energy, lost of entropy, geometry of directional interaction, local environment of interaction) Force Field SF • independant of training set • • strongly depends on ligand size force field accuracy, difficulty to set parameters no account of entropy Often includes empirical terms !

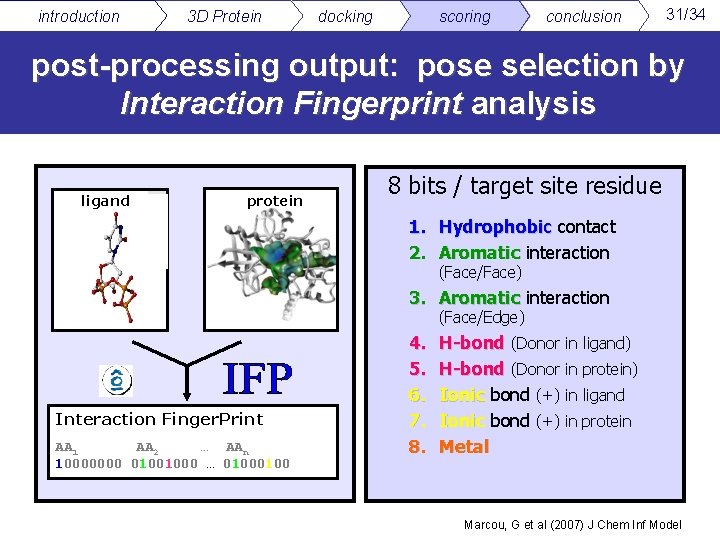
introduction 3 D Protein docking scoring conclusion 31/34 post-processing output: pose selection by Interaction Fingerprint analysis ligand protein 8 bits / target site residue 1. Hydrophobic contact 2. Aromatic interaction (Face/Face) 3. Aromatic interaction (Face/Edge) Interaction Finger. Print AA 1 AA 2 … AAn 10000000 01001000 … 0100 4. 5. 6. 7. 8. H-bond (Donor in ligand) H-bond (Donor in protein) Ionic bond (+) in ligand Ionic bond (+) in protein Metal Marcou, G et al (2007) J Chem Inf Model

32/34 conclusion What can we achieve? What remains to be improved?

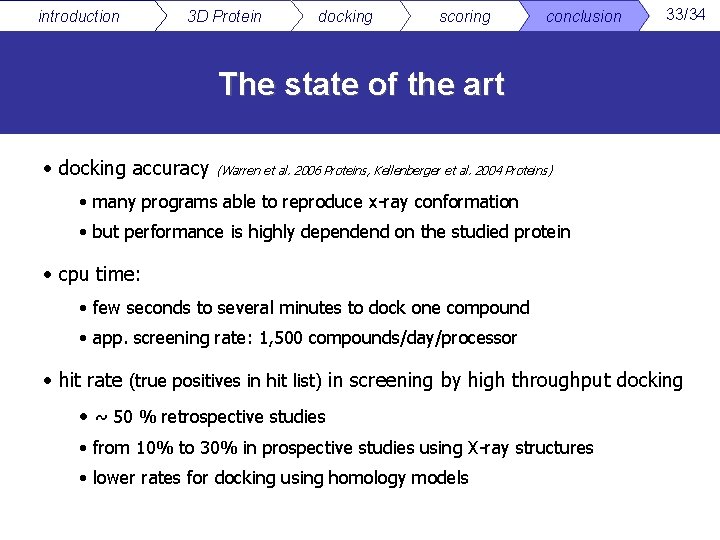
introduction 3 D Protein docking scoring conclusion 33/34 The state of the art • docking accuracy (Warren et al. 2006 Proteins, Kellenberger et al. 2004 Proteins) • many programs able to reproduce x-ray conformation • but performance is highly dependend on the studied protein • cpu time: • few seconds to several minutes to dock one compound • app. screening rate: 1, 500 compounds/day/processor • hit rate (true positives in hit list) in screening by high throughput docking • ~ 50 % retrospective studies • from 10% to 30% in prospective studies using X-ray structures • lower rates for docking using homology models

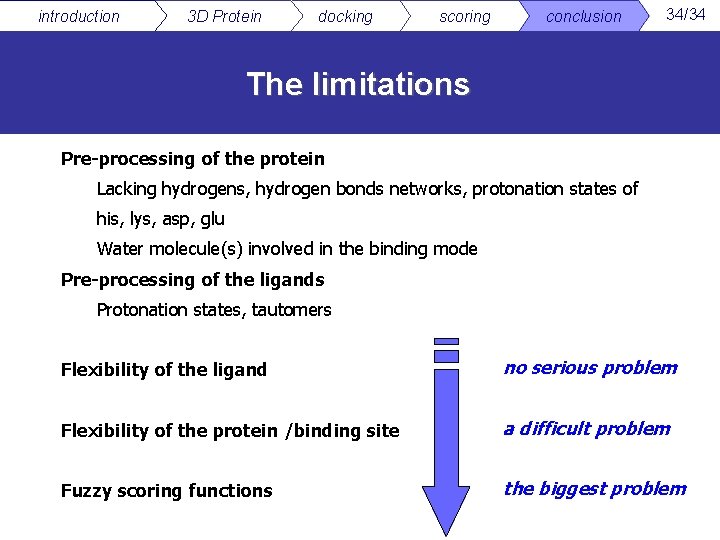
introduction 3 D Protein docking scoring conclusion 34/34 The limitations Pre-processing of the protein Lacking hydrogens, hydrogen bonds networks, protonation states of his, lys, asp, glu Water molecule(s) involved in the binding mode Pre-processing of the ligands Protonation states, tautomers Flexibility of the ligand no serious problem Flexibility of the protein /binding site a difficult problem Fuzzy scoring functions the biggest problem