Sigma Factors Transcriptional Regulation of P syringae TTSS



- Slides: 20

Sigma Factors & Transcriptional Regulation of P. syringae TTSS Alexander Wong

Presentation Outline RNApol holoenzyme n General properties of sigma factors n The alternative σ54 factor n Introduction to type III secretion system n Transcriptional regulation of Pseudomonas syringae TTSS n Conclusion n

The RNApol holoenzyme n Definition of holoenzyme ¨ Complete, working version of an enzyme ¨ cf. apoenzyme - missing specific cofactors that allow it to perform its job n Examples of cofactors ¨ common prosthetic groups (haem) or metal ions (magnesium) ¨ Dissociable protein subunits – sigma (σ) factor.

The RNApol holoenzyme n n n All multi-subunit RNA polymerases have 5 core subunits. Bacterial RNApol have additional σ subunit Has function in binding to promoter ¨ ¨ n n In bacteria, RNApol binds a promoter via σ In eukaryotes, RNApol binds via TF complex Bacterial RNApol is regulated purely by σ (initiation phase), but eukaryotic RNApol is regulated both by the TFs and by various gene regulatory proteins. Although promoters are similar, the bacterial promoter tends to be highly conserved.

Presentation Outline RNApol holoenzyme n General properties of sigma factors n The alternative σ54 factor n Introduction to type III secretion system n Transcriptional regulation of Pseudomonas syringae TTSS n Conclusion n

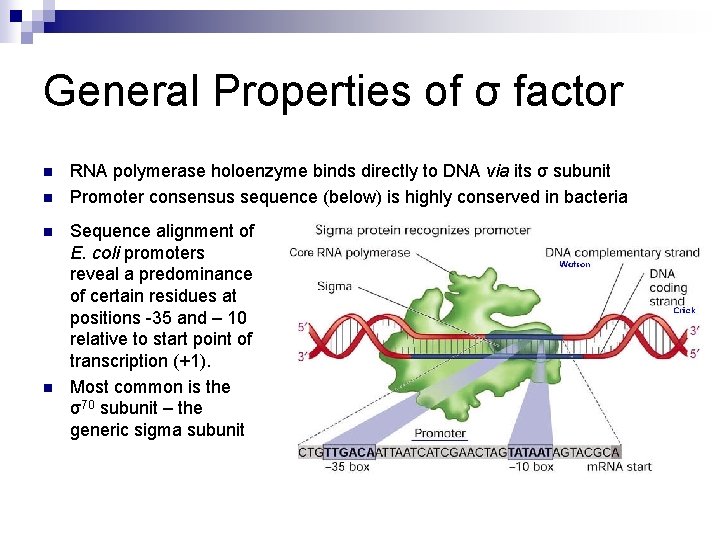
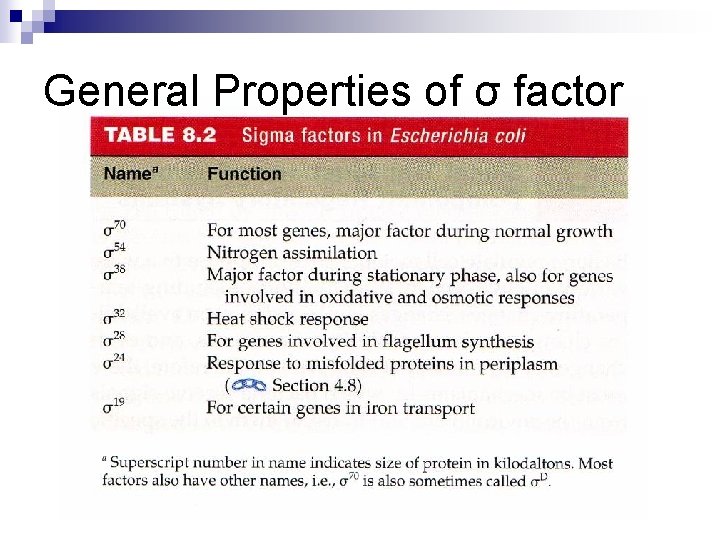
General Properties of σ factor n n RNA polymerase holoenzyme binds directly to DNA via its σ subunit Promoter consensus sequence (below) is highly conserved in bacteria Sequence alignment of E. coli promoters reveal a predominance of certain residues at positions -35 and – 10 relative to start point of transcription (+1). Most common is the σ70 subunit – the generic sigma subunit

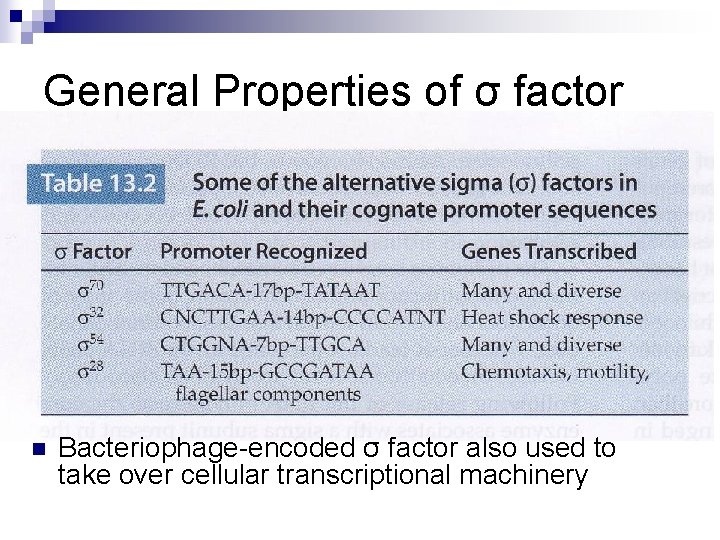
General Properties of σ factor

General Properties of σ factor n Bacteriophage-encoded σ factor also used to take over cellular transcriptional machinery

Presentation Outline RNApol holoenzyme n General properties of sigma factors n The alternative σ54 factor n Introduction to type III secretion system n Transcriptional regulation of Pseudomonas syringae TTSS n Conclusion n

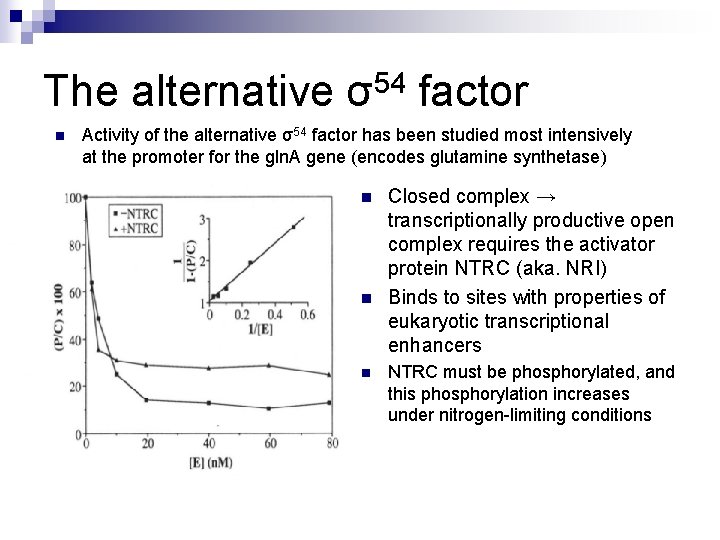
The alternative σ54 factor n n n Most alternative sigmas are related in sequence and structure to σ 70. 2 nd distinct type of σ called the σ54 family Differences between the σ families ¨ σ 54 family shares no sequence homology with the σ 70 family ¨ Whereas σ 70 holoenzymes carry out this process of open complex formation on their own, σ 54 holoenyzmes require both an enhancer and ATP to perform this process.

The alternative σ54 factor n Activity of the alternative σ54 factor has been studied most intensively at the promoter for the gln. A gene (encodes glutamine synthetase) n n n Closed complex → transcriptionally productive open complex requires the activator protein NTRC (aka. NRI) Binds to sites with properties of eukaryotic transcriptional enhancers NTRC must be phosphorylated, and this phosphorylation increases under nitrogen-limiting conditions

Presentation Outline RNApol holoenzyme n General properties of sigma factors n The alternative σ54 factor n Introduction to type III secretion system n Transcriptional regulation of Pseudomonas syringae TTSS n Conclusion n

Introduction to type III secretion system (TTSS) n n System with many names – PEC, injectisome, TTSS, TTS etc. Function to deliver bacterial proteins into target cells that then modulate host cell functions ¨ ¨ ¨ n n n Structural Translocation Effector proteins Structurally homologous to bacterial flagellum Genes usually clustered in mobile elements called pathogenicity islands (PAI) Significance of research in bacterial pathogenicity and potential medical application

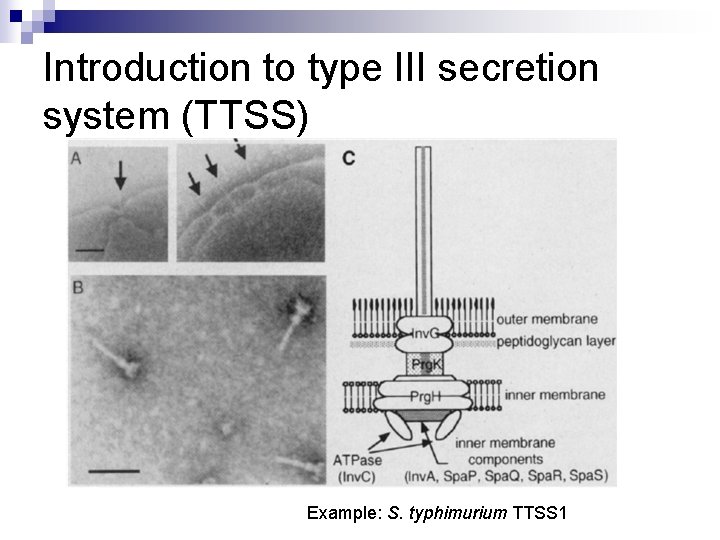
Introduction to type III secretion system (TTSS) Example: S. typhimurium TTSS 1

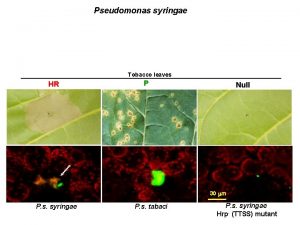
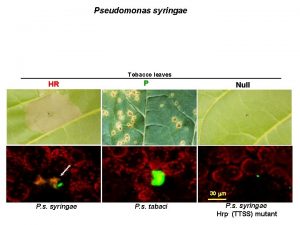
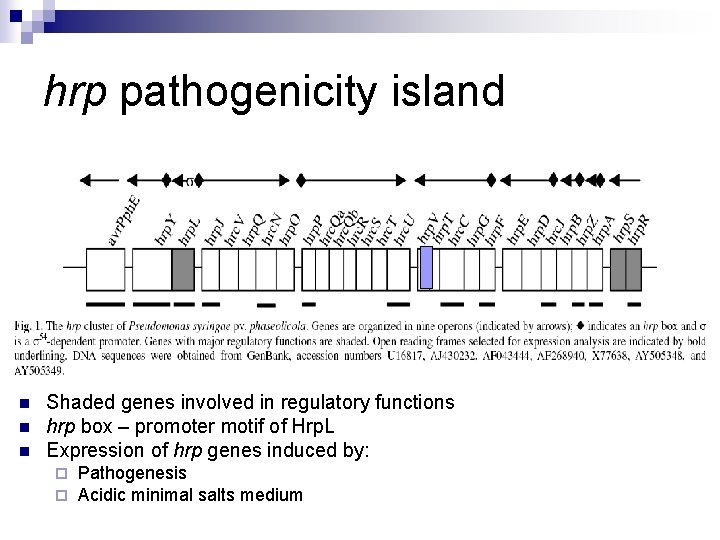
hrp pathogenicity island n n n Shaded genes involved in regulatory functions hrp box – promoter motif of Hrp. L Expression of hrp genes induced by: ¨ ¨ Pathogenesis Acidic minimal salts medium

Presentation Outline RNApol holoenzyme n General properties of sigma factors n The alternative σ54 factor n Introduction to type III secretion system n Transcriptional regulation of Pseudomonas syringae TTSS n Conclusion n

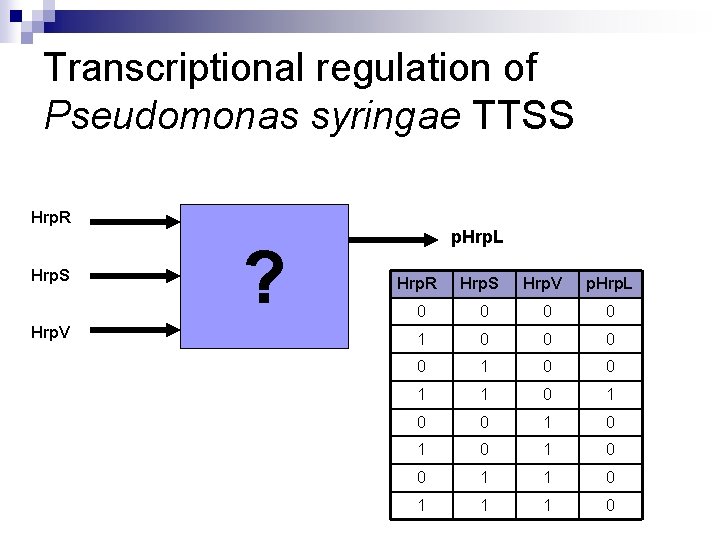
Transcriptional regulation of Pseudomonas syringae TTSS Hrp. R Hrp. S Hrp. V ? p. Hrp. L Hrp. R Hrp. S Hrp. V p. Hrp. L 0 0 0 0 1 1 0 0 1 0 1 0 0 1 1 1 0

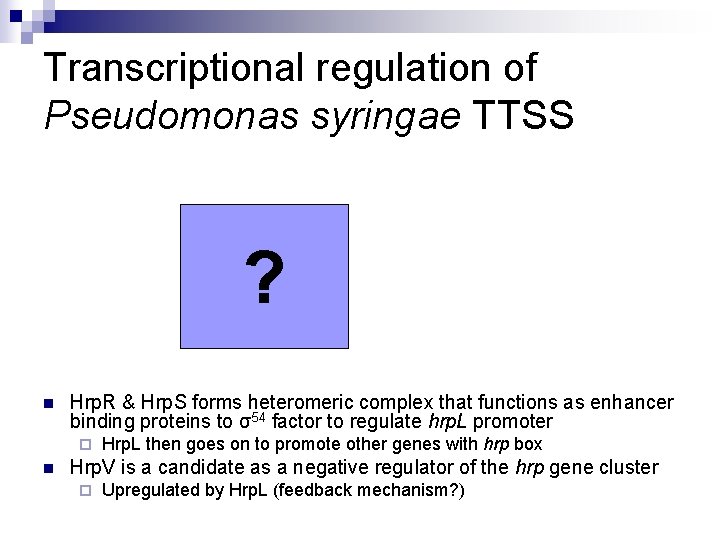
Transcriptional regulation of Pseudomonas syringae TTSS ? n Hrp. R & Hrp. S forms heteromeric complex that functions as enhancer binding proteins to σ54 factor to regulate hrp. L promoter ¨ n Hrp. L then goes on to promote other genes with hrp box Hrp. V is a candidate as a negative regulator of the hrp gene cluster ¨ Upregulated by Hrp. L (feedback mechanism? )

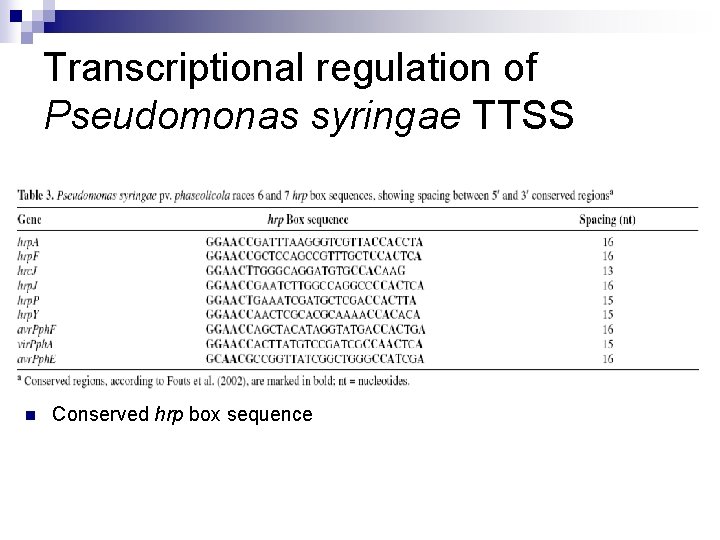
Transcriptional regulation of Pseudomonas syringae TTSS n Conserved hrp box sequence

Conclusion n n Candidate for i. GEM project? Considerations ¨ ¨ ¨ n Hrp. S could function as weak activator on its own (2. 5% activity) Extend usage of p. Hrp. L to Hrp. L and other effector proteins? ? Hrp. V needs a new promoter motif (regulated by Hrp. L) Noise reduction Requirement to strip gene cluster into individual components (other regulators involved) ¨ Protocol for optimal media conditions ¨ n Lab techniques ¨ ¨ RT-PCR Microarray and RT-PCR analysis done – what other data is required (particularly with negative regulation), and how much of the project can we call our own?
 Vcom white coat ceremony
Vcom white coat ceremony Vcom auburn white coat ceremony
Vcom auburn white coat ceremony 7-1 factors and greatest common factors
7-1 factors and greatest common factors Site vs. situation
Site vs. situation Abiotic vs biotic
Abiotic vs biotic Common factors of 8
Common factors of 8 Abiotic factor
Abiotic factor Site vs. situation
Site vs. situation Abiotic vs biotic factors
Abiotic vs biotic factors Greatest common factor powerpoint
Greatest common factor powerpoint Abiotic factors and biotic factors
Abiotic factors and biotic factors Regulation
Regulation Régulation
Régulation Site:slidetodoc.com
Site:slidetodoc.com Mandatory removal date army regulation
Mandatory removal date army regulation Human regulation
Human regulation Ar 670-1 religious accommodation
Ar 670-1 religious accommodation Malaysia elv regulation
Malaysia elv regulation Commercial banking regulation
Commercial banking regulation Foreign exchange transaction regulation korea
Foreign exchange transaction regulation korea Jtr 0505
Jtr 0505